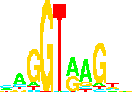The Software Needed for Making and Refining a Flexible Information Model
The Software Needed for Making and Refining a Flexible Information Model
by Nitasha Klar
Making A Sequence Logo
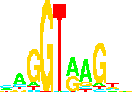
(Program names are in bold.)
catal
: cataloguer of delila libraries.
delila
: the librarian for sequence manipulation. It reads instructions
written in delila language from the inst file and puts the resulting output sequences
in a book.
alist
: reads the inst file to align the book generated by delila and creates a list,
one in color called the clist.
encode
: encodes a book of sequences into strings of integers and puts it in the encseq file.
comp
: determines the composition of a book.
rseq
: takes the encoded sequences from the encseq file and convers it to a
table of frequencies for each base at each aligned position. Rseq is calculated
and a weight matrix is generated which can be used to search for sites. The
output is stored in the rsdata file.
dalvec
: converts the rsdata file outputted by rseq into the symvec format
that the makelogo program can use.
ri
: rindividual is calculated for every site in the aligned book according to the
frequencies given in the rsdata file. ri outputs a ribl which is the
information content for each base at position "l". The ribl is used by scan
to scan over a different inst file of selected sites.
makelogo
: generates a sequence logo for a set of aligned sequences by reading
the rsdata file (output of dalvec) which is in the symvec format.
*run.logo
: runs all the logo making programs in one step, from delila through
makelogo.
Making A Walker

In addition to the above programs, the following programs are required for creating
walkers.
scan
: scans a book with a ribl weight matrix, outputted by the ri program,
and generates a vector.
lister
: lists the sequences of pieces in a book with translation.
genhis
: takes numerical data from a file and plots a histogram of those data. It
also calculates the min, max, mean and variance of the data.
discan
: compares the binding patterns of 2 different binding site models, it
selects sites that are within a certain range of each other and then
adds their individual information together and subtracts a distance
based distribution probability value to determine the new total
information.
diclean
: same as discan, except does not generate lister features.
Looking For a Stronger Alignment
markov
: generates a set of random dna sequences which have approximately the
same composition as the one in the composition file supplied to the
program.
embed
: embed an aligned set of DNA sequences into random sequences.
malign
: given a book of aligned sequences, this program searches for the alignment
of the sequences that has the lowest uncertainty, i.e. the highest
value of Rsequence.
malin
: makes delila instructions from the nth alignment of malign.
*sam
: a shell program that combines all the steps needed to malign in one, from
markov to malign.
The Refining Process:
Eliminating sites that have a negative information content.
mk.inst2
: a shell program that eliminates the negatives from the dsout/dcout file (outputted
from the discan/diclean programs, and makes delila
instruction files of the positive sites, using the makeinst program.
makeinst
: creates a delila instruction file using an input data file containing the
specified coordinates.
This page, created by
Nitasha Klar, was last modified
on March 24, 2000.


Schneider Lab
origin: 2000 Mar 24
updated: 2013 May 14


![]()

![]()