
Dissecting Dembski's "Complex Specified Information"
Thomas D. Schneider
This page is devoted to understanding
William A. Dembski's
concept "complex specified information" ("CSI").
My source is Dembski's book
No Free Lunch.
On this page
I investigate "CSI"
with respect to the
Ev model.
What is
"Complex"?
Page 111 says
The "complexity" in "specified complexity" is a measure of improbability.
This implies that it is a function of the probability of an object.
On page 127 Dembski introduces the information as
I(E) = def - log2 P(E),
where P(E) is the probability of event E.
Actually this is the surprisal introduced by Tribus
in his 1961 book on thermodynamics.
Be that as it may,
it is only a short step from this to Shannon's uncertainty
(which he cites on page 131)
and from there to Shannon's information measure,
so it is reasonable to use the significance of
Shannon's information
for determining complexity.
The main point about "Complexity" is that it be unlikely
that the observed bits occur. He is really concerned
with the significance of the measure.
So is the probability of the information gain in Ev sufficiently
low to say that the results are "complex"?
A test showed that in the standard Ev model the gain of 4 bits
is 12 standard deviations away from what one would expect by chance.
This is highly improbable.
I conclude that the Ev program produces Dembski's "complexity".
(Note: Having made this conclusion,
does not mean that I condone this or any
of Dembski's specialized terminology.
I think that 'complexity' is a vague term
with multiple conflicting definitions from many authors.
It should be avoided.
"Statistically significant" does the job just fine.)
What is
"Specified"?
This seems to be that the 'event' has a specific pattern.
In the book he runs around with a lot of vague definitions.
"Specification depends on the knowledge of subjects.
Is specification therefore subjective?
Yes."
Page 66
That means that if only if Dembski says something is specified, it is.
That's pretty useless of course.
Later on he tangles it up with
specificity,
which is a terrible term:
Biological specification always refers to function.
An organism is a functional system comprising many functional
subsystems.
In virtue of their function these
systems embody patterns that are objectively
given and can be identified independently of the systems that
embody them. Hence these systems are specified ...
(page 148)
So I'll take it that if one can make a
significant
sequence logo
from a set of sequences,
then that pattern is 'specified'.
Clearly Dembski would want these to fall under his roof
because they represent the natural binding patterns of proteins on DNA.
(Note: the concept of "specified"
is the point where Dembski injects
the intelligent agent that he later "discovers" to be design!
This makes the whole argument circular. Dembski wants "CSI"
rather than a precise measure such as Shannon information because
that gets the intelligent agent in. If he detects "CSI",
then
by his definition he automatically gets an intelligent agent.
The error is in presuming
a priori that the information must be generated
by an intelligent agent.)
What is
"Information"?
Dembski mentions Shannon uncertainty (p. 131)
without objection as the average of the 'surprisal'.
He accepts my use of the Rsequence measure too
(p. 212-218). So I will take information to be the Shannon
information.
How does this differ from the "complexity", which Dembski
also measures in bits?
On page 161 he states:
What turns specified information into
complex specified information is that the quantity
of information in the conceptual component is large.
In other words, "information" is in bits,
and
he measures
"complexity" in bits too but really means the significance.
Note that this confuses two separate issues.
A sequence logo made from a few sequences may have 10 bits
and not be very significant (large error bars)
while one made from thousands of sequences may have
very few bits
and be highly significant.
For example, position -25 of acceptor sites in our
1744 site model
(Stephens & Schneider, 1992)
has only
0.04 bits
but the probability of that occurring randomly
(as measured from the background noise)
is 1.5x10-8.
Another way to look at this is that the average of
1744 gave 0.04 bits, so the TOTAL across
all the measured genes is 1744x0.04 = 70 bits.
This number is Dembski's "complexity".
The disadvantage of the measure is that it depends
on sample size, but sometimes it is interesting to note
it.
In the standard Ev simulation, there are 16 binding
sites with 4 bits per site for a total of 64 bits
evolved in 704 generations to give a rate of about
1 bit every 11 generations.
The probability of this result by chance is
5x10-20.
(Note: On page 131 Dembski makes the same errors that most people
do these days, namely confounding information with uncertainty,
and then confusing uncertainty with entropy.
For example, on page 148 Dembski says that one can get 1000 bits
of "information" from 1000 coin flips. This is incorrect. He
should have said that one can get 1000 bits of uncertainty this way.
The uncertainty before flipping is 1 bit per flip.
At this point we have to consider how these random flips
are used, and there seem to be several possibilities.
One way is to count them as useful (e.g. in creating
a secret code), in which case
the uncertainty after is 0 bits per flip and the
information is the difference: 1 bit per flip.
The other way is to note that the results are not communication:
having received a coin state we would not have learned anything
(our uncertainty about all topics is the same)
and so
the uncertainty after is effectively still 1 bit per flip.
Again,
the information is the difference: 0 bits per flip.
In either case, it is important to consider the uncertainty of the
state before versus the state after.
If we just note that it is a stream of random flips then we
don't have information, we just have uncertainty.
See
the glossary definition of information for more details.
This error is pervasive in the literature,
but it is fatal for any serious project that has to deal
with real data.
The other error is to confound uncertainty
with entropy.
Dembski says they are mathematically identical.
That's wrong, they differ by an important multiplicative
constant. That is, they have different units.
If this is ignored, one is unable to proceed.
See
Information Is Not Entropy,
Information Is Not Uncertainty! for more details.)
What is
"Complex Specified Information"?
"Complex specified information is a souped up form of information."
(p. 142)
So a statistically significant (i.e.
"complex")
sequence logo, which has an
information
measured in bits
and a
"specified"
pattern shown by the graphic falls under the Specified Complexity
definition.
On page 111, Dembski says it has to have the "right" pattern.
Indeed, in many cases, the pattern is "right" in the sense of having the
Rsequence close to Rfrequency and
also in the sense that the binding site is bound
by the protein and this helps the organism to survive.
(Note: Aside from these interpretations,
"right" is too subjective and therefore vague, for general use.)
Two cases of
"Complex Specified Information"
Suppose we have these two sequence sets:
"Binding Site" Set 1
atacagacatttgctg
gactgggcctgaccag
tgggtagtattcttag
tgaccagagtttgaat
ctggaaagctgtcagc
gtttgaatatttaaaa
atttaaaaatggatgc
gccttaaggtggggac
gccttgaggttagagg
gttcggaagtgggtag
ttagaggagtgggcct
ggtggggactgggact
gagtgggccttaaggt
ttcttagccttgaggt
ttgctgacgttcggaa
gactgggactgggcct
|
"Binding Site" Set 2
tttgcgtaaataatcg
agtgactaaaatttac
gtggtgcgataattac
attgcatttaaaatga
aaaggccaaactttga
atgagttaagaaatga
gttgttcagtttttga
taagtatacagatcgt
aacgggcaataattgt
actgccagtttttcga
accgcgcaacattcaa
ttagcacactttttca
cacgatgaagaaacag
attggtcacattttat
ggcgattaaagaataa
aacggattaaaggtaa
|
which have these sequence logos:
|
|
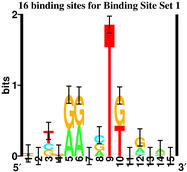
|
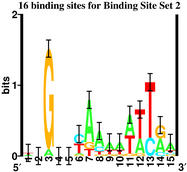
|
Although there is no information
about how I got these,
since both have a pattern
and both have nice significant
sequence logos, we have to conclude that both
have "Specified Complexity".
According to Dembski,
we must conclude that they both were generated
by an "intelligent designer".
Having made that unbiased conclusion,
I can now reveal that the logo on the left
was generated using the
ev program, while the one on the
right is the first few examples of
the left side of
natural
Fis sites
from our paper
Hengen et al Nucleic Acids Research, 25: 4994-5002 (1997).
The Ev model creates
"Complex Specified
Information"
So, though I find the "CSI" measure too vague to be sure,
for the sake of argument,
I tentatively conclude that the Ev program generates what Dembski
calls "complex specified information".
Evidently Dembski thought this in the summer of 2001
since at that time he tried to find
the source.
Where he asked, had Tom Schneider snuck in the CSI?
He suggested that it came from the SPECIAL RULE.
(Dembski's suggestion
turned out to be incorrect.)
Also,
in
No Free Lunch,
Dembski
asked where the "CSI" came from in Ev runs
(p. 212 and following).
So Ev creates "CSI".
According to Dembski, the existence of
"specified complexity" always implies an "intelligent" designer.
The question then is where, who or what is the designer?
Is it me, Tom Schneider?
No, that can't be right. I did set up the program - true - but
I didn't make the complexity. When I set it up the parameter file,
I only specified the size of the genome, the number of sites
and a few other items.
Well! The size of the genome and number of sites determines
the information Rfrequency! But wait. At the first generation the
sequence is RANDOM and the information content Rsequence is ZERO.
So I didn't make the "complex specified information",
as measured by Rsequence
after 2000 generations.
It must be created by Ev.
Nor did I make the 'right' ("specified")
pattern match the weight matrix gene to the binding sites.
Only
after
I took my hands off the parameter file was
the random sequence generated and the evolutionary process started.
When I was involved there was no correlation between the two.
Ev must have made the correlation.
One could vary the parameter file using a random number
generator. Given what we know about how ev works,
unless one hits an extreme (e.g. binding site width
too small to contain the required information) the evolution
should show information gain in every case.
So even my selection of parameters is irrelevant.
If you don't believe me, go
get the program
and
do the experiment yourself!
(Note: You can now
run the Evj java version
on your own computer.)
Ev uses a pseudorandom number generator, so one might
complain that the results are "preordained".
If you feel this way, I invite you to substitute into the
program any good generator.
A good generator would give a flat distribution between 0 and
1 and not repeat for at least thousands of calls.
You could even use the
HotBits radioactive source
(http://www.fourmilab.ch/hotbits/) and remove any
lingering doubt!
Having run the program with different generators in various
ways, it is clear that this cannot be the source of the "CSI".
(Note: Dembski would probably accept the output of HotBits
since he mentions radioactive emission as
a generator of "pure chance" on page 149.)
Was it, as Dembski suggests in
No Free Lunch (page 217),
that I programmed Ev to make selections based on mistakes?
Again, no.
Ev makes those selections independently of me.
Fortunately I do not need to sit there
providing the
selection
at every step!
The decisions are made given the current state,
which in turn depends on the mutations which come from the
random number generator.
I personally never even see those decisions. Of course
since it is a program, one could watch them if wanted to by using the
appropriate printouts.
By contrast,
a major weakness of Dawkin's biomorphs
(described in
"The Blind Watchmaker",
W. W. Norton & Co.,
New York,
1986)
was that a person had to make a decision at every step
of the process by selecting a shape that they liked.
The
METHINKS IT IS LIKE A WEASEL
example was better, but obviously had a specific final pattern.
Ev, in contrast, is not guided at each step and has no specific
final pattern. Rfrequency is only by the size
of the genome and number of sites, but these may be varied
over a large range and there still is an information gain
during the Ev run. Watch the
ev movies
to see that the
pattern shifts over time after the Rsequence reaches
Rfrequency.
It is the selections made by the Ev program
that separates organisms
with lower information content from those that have higher
information content.
If no selections are made, then
it is straightforward to demonstrate
that there is no information gain.
So the Ev program itself must be the intelligent designer.
How does Ev do that? By mutation (non directed, can't be intelligent)
replication (non-directed, can't be intelligent) and selection (ahh ha!).
So the 'intelligent' component must be selection.
But this is exactly what happens in nature, as far as we can tell.
Thousands of papers give evidence that mutation occurs.
For examples of observed human mutations
in splice junction binding sites,
see
ABCR
and
RFS.
These mutations cause diseases and we know that the people who have them
do not survive as well as others, so there is selection
for their binding sites to be good.
We also know that people replicate.
All the components that Ev has are the same as in nature.
So what's going on?
Living things themselves
create "specified complexity"
via environmental selections and mutations.
Living things and their environment are the "intelligent designer"!
So what was Dembski's mistake?
It was that he proposed that the design by necessity had
to come from outside the living things, whereas it
comes from within them
and between the organism and its environment!
Normally this is called evolution by natural selection.
Coda
Dembski claimed that he could prove his thesis mathematically:
In this section I will present an in-principle mathematical argument for
why natural causes are incapable of generating complex
specified information." (page 150)
He shows that pure random chance cannot create information,
and
he shows how a simple smooth function
(such as y = x2)
cannot gain information.
(Information could be lost by a function that cannot be mapped
back uniquely: y = sine(x).)
He concludes that there must be a designer to obtain CSI.
However,
natural selection has a
branching
mapping from one to many
(replication) followed by
pruning
mapping of the many back down
to a few (selection). These increasing and
reductional mappings were not modeled
by Dembski. In other words, Dembski "forgot" to model birth and death!
It is amazing to see him spin pages and pages of math which are
irrelevant because of these "oversights".
Dembski's entire book, No Free Lunch,
relies on this flawed argument,
so the entire thesis of the book collapses.
Discussion


Schneider Lab
origin: 2002 March 5
updated: version = 1.12 of specified.complexity.html 2008 Dec 01




