 ev: Evolution of Biological Information,
ev: Evolution of Biological Information,
Evj is an evolutionary model that runs inside your computer using the Java language. This page is a beginner's guide to experimenting with Evj. For more information you can read the original scientific paper, "Evolution of Biological Information".
Evj models the evolution of genetic control systems. In all living organisms, genes are parts of the DNA that (usually!) code for proteins. The proteins do all kinds of things for the cell, such as controlling whether other proteins are made or not. The regulatory proteins do this by binding to the DNA and turning on or off the genes of the other proteins. The classical example of gene regulation is the Lac Operon. Here are some pointers to read more about it:
| Click Here to Start the Model |
| Click Run |
| Click Pause |
| Controls and Displays |
|
Genome A C G T . . . |
| red | binding site, not functional |
| green | binding site, functional |
| yellow | binding at the wrong place |
| blue | gene weight, positive |
| cyan | gene weight, negative |
| Mutate each creature by changing its genome randomly. For example, the program might change the letter at position 82 from an A to a T. Mutation happens everywhere in the genome, both in the gene, in the binding sites, and in the spaces between. The location is random and the change is random. |
Note that mutations often occur during DNA replication
in nature when the wrong base is inserted.
Another mechanism is DNA damage, which can cause
the wrong complementary base to be inserted.
Mutations happen naturally, often from radioactive and other compounds in the environment, but you can speed up the process. If you smoke cigarettes, you put chemicals into your body that cause mutations in your cell's DNA. Eventually, some of your cells might lose their genetic growth controls, and will start to grow wildly. They could take over your lungs and kill you. This disease is cancer. |
|
Evaluate
each creature: count the number of mistakes
each one makes.
This is the first step of selection. | Some mutated lungs cells will not do well, but others might be able to grow without normal controls. Most mutations wreck controls; however, on occasion the random changes will improve a control protein binding site. It's easier to wreck your (old fashioned!) television or radio by hitting it than to make it work, but on occasion you might be lucky. Of course electronic equipment is complex and hitting is blunt, so unless you jiggle a disconnected wire into place, hitting it is not likely to help. In contrast, binding sites are so simple that changing them may often make the protein bind better. |
|
Sort
the creatures by their mistakes.
This is the second step of selection. | |
|
Kill
half of the creatures,
the ones that make the most mistakes.
This is the final step of selection. | You probably kill some of your lung cells by smoking. (Searching PubMed for 'lung cells death smoke' led to Study of the mechanisms of cigarette smoke gas phase cytotoxicity. Anticancer Res. 2003 May-Jun;23(3A):2185-90. Piperi C, Pouli AE, Katerelos NA, Hatzinikolaou DG, Stavridou A, Psallidopoulos MC..) |
| Replicate (make another copy of) the creatures that make the least mistakes. |
Lung cells will grow back if they survive the
mutagens in the cigarettes.
Cells that lose their genetic growth controls
could grow much faster and then you would get
cancer.
That is, cancer is caused by evolution by natural selection
of cells inside your body.
It's your choice whether you want to help it along or not
by increasing your mutation rate by smoking.
Likewise,
tanning exposes you to
UV radiation from the sun that can mutate your DNA,
and eating fried foods can too.
|
| Questions |
| Experiment Number 1: Flying through Evolution. |
| Click Restart |
| Click Run |
| Crank up the Speed |
| Experiment Number 2: The Effects of Selection. |
| Click on the check box next to the word Selection. |
|
Experiment Number 3: Flickering Bases - Sequence Conservation versus Neutral Drift. |
Here is a screenshot for reference. This is the results of a standard run to 10,000 generations on my machines. You should be able to get the same result just by repeating Experiment 1 and letting it go until it stops.
Now take a look at the gene, which is marked with Blue and cyan boxes. Each box is marked with a number and a base 0A, 0C, 0T, etc, followed by a number called a 'weight':| 0 | 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|---|
| A | -159 | -386 | -148 | -326 | -21 | +363 |
| C | -450 | +193 | +127 | +341 | -71 | -178 |
| G | -266 | -28 | -52 | -10 | -481 | -149 |
| T | -151 | -342 | +510 | -252 | -187 | -178 |
C is under position 0 of the table,Then we pick the number from the row corresponding to the letter:
G is under position 1,
T is under position 2,
C is under position 3,
T is under position 4,
A is under position 5.
C is -450,Finally, we add these together to get +549. This is the number written on the site at position +190. If you look at the genome, you will see that there is one more number at the end of the gene called 'th', which stands for 'threshold'. In this case, the threshold is +300. If the sum of the weights is bigger than the threshold, then the protein model has found a binding site. For the site at +190, this is true, so the site is marked with a green box.
G is -28,
T is +510,
C is +341,
T is -187,
A is +363,
What is Neutral Drift?
Kimura was the person who introduced the idea of neutral drift.
These are changes to the genome that have little effect on survival.
Here's one of his papers:
KIMURA M, CROW JF. THE NUMBER OF ALLELES THAT CAN BE MAINTAINED IN A FINITE POPULATION. Genetics. 1964 Apr;49:725-38. PMID: 14156929 Of course back in the 1960s they didn't have lots of sequence data so he made mathematical models. In contrast, the Ev model has an explicit genome and actual functions. A Google search for Kimura neutral gives a page by Gert Korthof who says: I included this work of Kimura to show that a critique of Darwinism is possible, without being ridiculed or ignored by the scientific community.Intelligent design is being ridiculed and ignored because it is bad science. The ideas don't hold up to careful scrutiny and when the ideas are disproven, it is not acknowledged. Kimura's idea did hold up to scrutiny, and we can see it in the Ev program when running full blast. Just watch the regions outside a binding site flicker! |
| Experiment Number 4: Understanding Aligned Sites. |
By now you have surely noticed the jumping piles of letters in the control and display region. Sorry to make you wait for an explanation about them!
Suppose we list all of the sites vertically:

How many t's are at position 2 in the sites? (Answer)
| Experiment Number 5: Understanding Sequence Logos. |
How can we represent this complex pattern of letters? The sequence logo can do this:
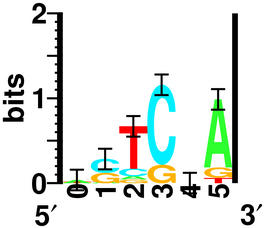
The positions of the logo correspond to the positions in the sites. So let's look at position 2. At that position is a stack of letters, with T on the top because it is the most frequent base at that position. Below the T is a C because that is the next most frequent base. Under that (if you look closely!) you will find an A (in green) and a G (in orange). The rule is that the height of each letter is proportional to the frequency of that base at that position in the site.
What determines the height of the stack of letters? The answer is that we can measure how conserved the letters are in bits of information. This is a longer story than can be fully explained here, but here is a pointer to get you started with the definition of a bit.
So, to summarize, the sequence logo shows you in a compact graphical form which parts of the binding site are conserved and precisely by how much.
Restart the simulation, Set the "Cycles to run" to 1000 and and click on Run. Watch the last base of the site in both the sequence logo and the genome. What happens? (Answer)
| Experiment Number 5: Watching Evolution with Sequence Logos. |
Restart the model, crank up the "Cycles to run" to at least 100,000, and click Run. Once a sequence logo has emerged, turn off selection. (The selection box may not respond. In that case, Pause, click the selection box and Run.) What happens to the logo? (Answer) What happens to the logo when you turn selection on again? (Answer)
| Experiment Number 6: How binding sites evolve: Rsequence and Rfrequency. |
The heights of the sequence logo stacks are in bits of information. It turns out that these are related to the size of the genome and the number of sites. To learn more about this, you can read the article The Nitty Gritty Bit.
This page is:
https://alum.mit.edu/www/toms/paper/ev/evj/evj-guide.html.
A tinyurl
for this page is
http://tinyurl.com/evolution-in-a-nutshell.
You can preview the tinyurl with
http://preview.tinyurl.com/evolution-in-a-nutshell
Acknowledgments. Thanks to Pete Lemkin and Adam Diehl for useful comments on this page.
Problems? Comments? Please email me, Tom Schneider, at toms@alum.mit.edu.
![]()

Schneider Lab
origin: 2005 Jun 3
updated: 2012 Jan 01 version = 1.47 of evj-guide.html
![]()