 ev: Evolution of Biological Information:
ev: Evolution of Biological Information:
Phenomena are recorded as data (information) when the state of a device (including things like rhodopsin in the eye and CCDs) associated with a living organism is changed by the phenomena.I'm only a few thousand years behind in this thought:
"By convention there is colour, by convention sweetness, by convention bitterness, but in reality there are atoms and space."
--- Democritus - 400 B.C.E.
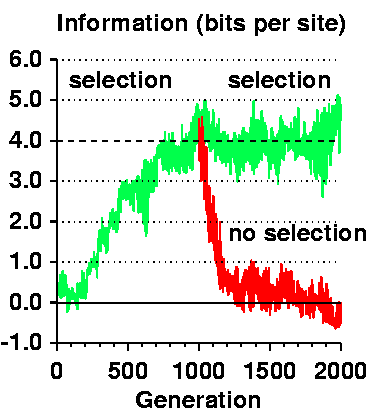
Rfrequency = - log 2 (γ / G)Because Rfrequency is a function of the size of the genome (the number of potential binding sites is G) and the number of sites (γ), it is fixed when the model begins and (usually) is not changed during an evolution run. So it doesn't teach us about how Rsequence changes. However, Rsequence does evolve towards Rfrequency as you can see in Figure 2b of the Ev paper and in the figure to the right. The dashed line shows Rfrequency, the green curve shows Rsequence.
|
are necessary and sufficient for information gain to occur. This process is called evolution. |

| Statistical decision |
|
|||||
| Reject Ho = site found |
Type I error | Correct | ||||
| Do not Reject Ho = site not found |
Correct | Type II error | ||||
green for sites found in the right place,
red for sites missed from the right place,
yellow for sites found in the wrong place
but
blue is normally not displayed.
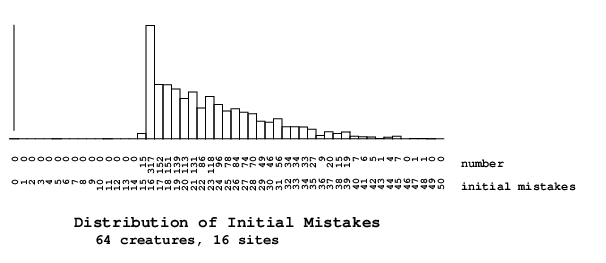
I ran the program with a few different seeds, and the best organism is at the first step already in a great shape, with only around 20 mistakes. I think that is not a reasonable starting state for the population; the best organism at the first step should have at least about 200 mistakes, if not be even closer to the maximum number of mistakes. (Unfortunately, I cannot modify the threshold to deal with that, and I am not going to try more seeds either, since it does not appear to go anywhere far from those values.)You didn't say what your parameters were, but suppose that you have 16 sites and 64 organisms as in the standard java run. Sorting gives the best organism, of course, so right away you have a strong bias. Why 20? I guess that this is most easily "accomplished" by having a weight matrix that does not recognize ANYTHING, or has little recognition capability. If it didn't recognize anything there would be exactly 16 mistakes. This could happen by having a very high initial threshold. If it accidently recognized 4 more sites in the wrong locations that would account for your 20. This is a hypothesis and so you can test it by looking closely at the organism that has that situation. I do agree it is a somewhat curious effect. Would it happen in nature? Sure. All that has to happen is a recognition protein is duplicated (apparently a common occurance since we see lots of nearly identical genes in various organisms and the recombination mechanism for doing this is pretty well understood). Then one copy diverges so that it doesn't recognize much at all on the DNA. As it then starts to locate a few spots, if it matches, WHOSH selection takes over and it locks on. This effect occurs in Ev too of course.
![]()

Schneider Lab
origin: 2005 May 24
updated: 2013 May 08
![]()