
flexprom: Anatomy of Escherichia coli sigma70 promoters
|
|


|

|
@article{Shultzaberger.Schneider-flexprom2007,
author = "R. K. Shultzaberger
and Zehua Chen
and Karen A. Lewis
and T. D. Schneider",
title = "{Anatomy of \emph{Escherichia coli} $\sigma^{70}$ promoters}",
journal = "Nucleic Acids Res.",
volume = "35",
pages = "771--788",
pmid = "17189297",
pmcid = "PMC1807945",
note = "\htmladdnormallink
{https://alum.mit.edu/www/toms/papers/f{l}exprom/}
{https://alum.mit.edu/www/toms/papers/f{l}exprom/}",
\htmladdnormallink
{http://dx.doi.org/10.1093/nar/gkl956}
{http://dx.doi.org/10.1093/nar/gkl956}",
year = "2007"}
In this paper we predicted that base +4 (conventional base number -7) flips out of the DNA to initiate transcription. This was confirmed by two papers:
@article{Feklistov.Darst2011,
author = "A. Feklistov
and S. A. Darst",
title = "{Structural basis for promoter -10 element recognition by
the bacterial RNA polymerase sigma subunit}",
journal = "Cell",
volume = "147",
pages = "1257--1269",
pmid = "22136875",
pmcid = "PMC3245737",
year = "2011"}
(see Figure 1)
@article{Zhang.Ebright2012,
author = "Y. Zhang
and Y. Feng
and S. Chatterjee
and S. Tuske
and M. X. Ho
and E. Arnold
and R. H. Ebright",
title = "{Structural basis of transcription initiation}",
journal = "Science",
volume = "338",
pages = "1076--1080",
pmid = "23086998",
year = "2012"}
(see reference 11)
See also:
@article{Liu.Kornberg2011,
author = "X. Liu
and D. A. Bushnell
and R. D. Kornberg",
title = "{Lock and key to transcription: sigma-DNA interaction}",
journal = "Cell",
volume = "147",
pages = "1218--1219",
pmid = "22153066",
year = "2011"}
@article{Darst.Gross2014,
author = "S. A. Darst
and A. Feklistov
and C. A. Gross",
title = "{Promoter melting by an alternative sigma, one base at a
time}",
journal = "Nat Struct Mol Biol",
volume = "21",
pages = "350--351",
pmid = "24699085",
comment = "reference 9 cites Shultzaberger.Schneider-flexprom2007",
year = "2014"}
Start coordinates for sigma70 in E. coli accession U00096:
sigma70_start_coords.txt
Components for the sigma70 model
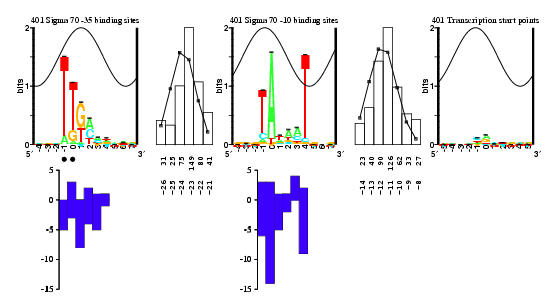
prom.eps is the original Figure 1 of the paper.
Logo for the sigma70 model with conventional numbering.
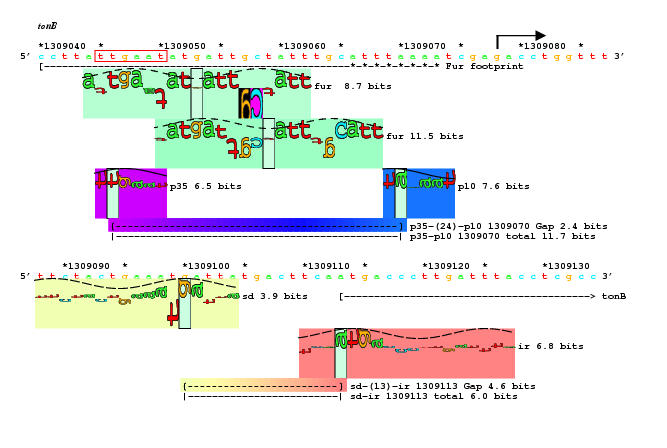
We used this structure given by Campbell.Darst2002 to determine how the sigma 70 -35 binds to DNA.
@article{Campbell.Darst2002,
author = "E. A. Campbell
and O. Muzzin
and M. Chlenov
and J. L. Sun
and C. A. Olson
and O. Weinman
and M. L. Trester-Zedlitz
and S. A. Darst",
title = "{Structure of the bacterial RNA polymerase promoter
specificity $\sigma$ subunit}",
journal = "Mol Cell",
volume = "9",
pages = "527--539",
pmid = "11931761",
year = "2002"}
These files show how the 4.2 part of the sigma protein binds into the -35 region of a promoter. Run them as:
rasmolscript 1KU7.pdb -script 1KU7.ras rasmolscript 1KU7-nowater.pdb -script 1KU7-nowater.ras
See also the companion paper, Anatomy of Escherichia coli Ribosome Binding Sites
The Delila Server allows you to try the model on the E. coli genome.
Other pointers:
![]()

Schneider Lab
origin: 2004 May 13
updated: 2017 Jul 14
![]()