Sequence Logos: A New Way to Display Consensus Sequences
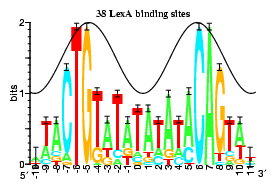
Sequence logo of LexA binding sites
|

|
Citations to sequence logo papers per year.
blue:
logo paper (from 1991 to present)
green:
weblogo paper (from 2004 to present)
red:
both papers (from 1991 to present, top curve)
The data are from the
ISI Web of Science's
Cited Reference Search.
Total growth is exponential to at least 2010,
but citations to the original logo paper
leveled out starting in 2004 when Weblogo became available.
The total may be leveling out by 2015.
|
|
@article{Schneider.Stephens1990,
author = "T. D. Schneider
and R. M. Stephens",
title = "Sequence Logos: A New Way to Display Consensus Sequences",
journal = "Nucleic Acids Res.",
volume = "18",
pages = "6097--6100",
pmid = "2172928",
pmcid = "PMC332411",
note = "\url{http://dx.doi.org/10.1093/nar/18.20.6097},
\url{https://alum.mit.edu/www/toms/papers/logopaper/}",
year = "1990"}
The 50 Most-Frequently-Cited Articles
in Nucl. Acids Res.
-- updated monthly
Historical Note:
We devised the title of this paper before we realized
that a sequence logo is more than merely a way
to display consensus sequences.
Of course it is that, since
one can read the strict consensus from the top letter
at each position of the logo.
But in addition one gets the frequencies and
the total sequence conservation, so logos are more
that just a display of consensus.
It takes time to begin to think in a new way!
Programs
This is a very
incomplete list of sequence logo servers and extensions to logos.
Citations
Questions
Contest!
2015 Jan 28: Google Ngram use of 'sequence logo'
This graph shows the percentage of time that
the phrase 'sequence logo' appeared in books in different years using the
Google Books Ngram Viewer.


Schneider Lab
origin: 1996 Feb 09
updated:
2024 Jul 09: update reference



