Splice Junction Analysis Service
 Jim Ellis
(DHHS/NIH/OD/ORS/DBEPS,
jrellis@helix.nih.gov,
voice: 301-496-4472, fax: 301-480-1242,
Bldg. 13, Rm 3E-49,
NIH directory entry)
has set up
Tom Schneider's
Delila programs
to do
splice junction analyses
for scientists at NIH
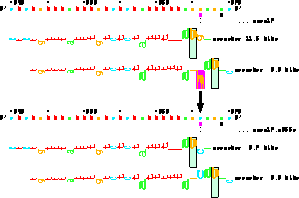
(P. K. Rogan, B. M. Faux and T. D. Schneider,
Information analysis of human splice site mutations
Human Mutation
12(3):
153-171 (1998).)
Published or
unpublished sequence data can be used but it is most efficient to start with
GenBank flat file format. Mutations are expressed in a precise computer
language called Delila.
Jim Ellis
(DHHS/NIH/OD/ORS/DBEPS,
jrellis@helix.nih.gov,
voice: 301-496-4472, fax: 301-480-1242,
Bldg. 13, Rm 3E-49,
NIH directory entry)
has set up
Tom Schneider's
Delila programs
to do
splice junction analyses
for scientists at NIH
(P. K. Rogan, B. M. Faux and T. D. Schneider,
Information analysis of human splice site mutations
Human Mutation
12(3):
153-171 (1998).)
Published or
unpublished sequence data can be used but it is most efficient to start with
GenBank flat file format. Mutations are expressed in a precise computer
language called Delila.
Dr. Ellis also does analyses as collaborations
with people outside NIH if he has time.
Dr. Ellis can begin the analysis if he has:
- The sequence, in decreasing order of preference as:
- A GenBank accession number with exons identified.
- A sequence in GenBank flat file format with exons identified
- The sequence with capital letters for primers (generally shorter
than 25 bp) and exons (generally longer than 25bp).
(By putting the sequence into the simple format required by the
mkdb
program, the process will be faster.)
and
- The exact sequence change(s).
Sequences and delila instructions
can be given to Jim on a floppy disk or by email.
Tips:
-
If you would like to speed this process, you can
write out the sequence
changes in
Delila
instructions.
Other mutation specifications are acceptable, but the process may be a bit less
efficient. Users do not have to learn Delila to use the service or obtain
results. Please call or e-mail if you have any questions.
-
Dr. Peter Rogan has suggested that sometimes sequence changes in the interior
of an exon will have splicing effects, so all available changes may be
interesting and should be looked at.
Changes that we think might not be
splice junction changes - because they are far from the known junctions -
might indeed play a roll in splicing.
For one thing, they could be a site
used in a processing event for which we haven't captured a cDNA yet. Another
possibility is the generation of cryptic sites. So our current thinking is
that we should look at all known mutations that affect the RNA.
Fortunately
a major revision of Delila (in 1999) allows one to make mutations quickly.
This allowed us to go through about 100 mutations in 2 days.
After you have
looked at your most interesting changes, you might consider going through all
of the known ones just to see if something interesting turns up.
- At least
26 bases
of intron are required for analysis
of acceptor sites
(-25 to +2 with
coordinate
zero being
the first base on the intron side of the junction)
and
7 bases
of intron are required for analysis
of donor sites
(-3 to +6 with coordinate zero being
the first base on the intron side of the junction).
More sequence will allow checking for cryptic sites. In general it is
best to provide all of the known sequence.
Publications
-
Splice mutations in NF2,
Kluwe Lan, Heinrich B, Ellis JR, Schneider TD, MacCollin M, Mautner V-F.
Abstract
presented at the 8th European Neurofibromatosis Meeting; September
23-26, 1999; Ulm, Germany.
-
Molecular Genetics and Metabolism 73 (4), August 2001,
333-339 (doi:10.1006/mgme.2001.3209)
Is the SLC7A10 Gene on Chromosome 19 a Candidate Locus for Cystinuria?
Daniel Leclerc, Qing Wu, James R. Ellis, Paul Goodyer, Rima Rozen
PubMed Abstract
-
Leclerc, D., M. Boutros, D. Suh, Q. Wu, M. Palacin, J. R. Ellis, P. Goodyer, and
R. Rozen. "SLC7A9 mutations in all three cystinuria subtypes", Kidney Int,
62(5): pp. 1550-1559 (2002).
PubMed Abstract
-
Hellerud, C., A. Burlina, C. Gabelli, J. R. Ellis, P.-G. Nyholm, and S.
Lindstedt. "Glycerol Metabolism and Determination of Triglycerides - Clinical,
Biochemical, and Molecular Findings in Six Subjects". Clinical Chemistry and
Laboratory Medicine, Vol. 41 (2003), pp. 46-55.
PubMed Abstract
-
Hellerud, C., M. Adamowicz, D. Jurkiewicz, J. Taybert, J. Kubalska, E.
Ciara, E. Popowska, J. R. Ellis, S. Lindstedt and E. Pronicka.
"Clinical heterogeneity and
molecular findings in five Polish patients with glycerol kinase deficiency:
investigation of two splice site mutations with computerized splice junction
analysis and Xp21 gene-specific mRNA analysis".
Molecular Genetics and Metabolism,
79(3) (2003), pp. 149-159.
PubMed Abstract
-
J. E. Cleaver, C. Collins, J. Ellis and S. Volik,
"Genome sequence and splice site analysis of low-fidelity DNA
polymerases H and I involved in replication of damaged DNA, Genomics",
Volume 82, Issue 5, November 2003, Pages 561-570.
PubMed Abstract;
at
www.sciencedirect.com
-
Goldin E, Stahl S, Cooney AM, Kaneski CR, Gupta S, Brady RO, Ellis JR,
Schiffmann R.
Transfer of a mitochondrial DNA fragment to MCOLN1 causes an inherited
case of mucolipidosis IV.
Hum Mutat. 2004 Dec;24(6):460-5.
PubMed Abstract.
-
Genes Chromosomes Cancer. 2011 Aug;50(8):571-84.
Effects of splicing mutations on NF2-transcripts: Transcript analysis
and information theoretic predictions.
Ellis JR Jr, Heinrich B, Mautner VF, Kluwe L.
PubMed Abstract.


Schneider Lab
origin: 1999 Apr 30
updated: 2011 Jun 27

 Jim Ellis
(DHHS/NIH/OD/ORS/DBEPS,
jrellis@helix.nih.gov,
voice: 301-496-4472, fax: 301-480-1242,
Bldg. 13, Rm 3E-49,
NIH directory entry)
has set up
Tom Schneider's
Delila programs
to do
splice junction analyses
for scientists at NIH
(P. K. Rogan, B. M. Faux and T. D. Schneider,
Information analysis of human splice site mutations
Human Mutation
12(3):
153-171 (1998).)
Published or
unpublished sequence data can be used but it is most efficient to start with
GenBank flat file format. Mutations are expressed in a precise computer
language called Delila.
Jim Ellis
(DHHS/NIH/OD/ORS/DBEPS,
jrellis@helix.nih.gov,
voice: 301-496-4472, fax: 301-480-1242,
Bldg. 13, Rm 3E-49,
NIH directory entry)
has set up
Tom Schneider's
Delila programs
to do
splice junction analyses
for scientists at NIH
(P. K. Rogan, B. M. Faux and T. D. Schneider,
Information analysis of human splice site mutations
Human Mutation
12(3):
153-171 (1998).)
Published or
unpublished sequence data can be used but it is most efficient to start with
GenBank flat file format. Mutations are expressed in a precise computer
language called Delila.

