Introduction to Tom Schneider's Laboratory
In our laboratory we use Claude Shannon's information theory, computers (Pascal and PostScript graphics on Unix computers) and genetic engineering (protein and DNA gels, cloning, sequencing and magnetic bead technology) to study genetic control patterns on DNA and RNA. We use computers to analyze published sequence data and we display the results like this:
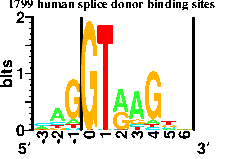
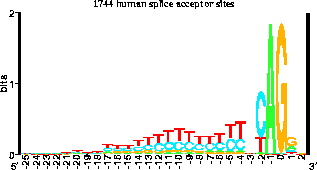
This is a ``sequence logo'' that I invented with my first high school student, Mike Stephens (Linganore High School). After RNA is transcribed from DNA in the cell, pieces are cut out in a process called splicing. The cell needs to know exactly where to cut the RNA, and these points are called donor and acceptor splice sites. Mike collected the sequences of 1744 human splice acceptors and then we had the computer generate this picture of them. The height of each stack of letters is in bits, and shows how ``important'' or how conserved that position is. The height of each genetic letter (A, C, G or T) is proportional to its frequency in the acceptors. At a single glance you can tell what the acceptors are like. We published the results in the Journal of Molecular Biology (228: 1124-1136, 1992). Another project by Nate Herman (Frederick High School) led us to predict the existence of several DNA binding proteins (Journal of Bacteriology 174: 3558-3560, 1992). In our lab, students have usually done work on the computer, but some projects are primarily hands-on lab bench work.
This is a brief 9 page introduction to molecular information theory.
This is a longer 37 page review of molecular machine theory.
Just in case you are confused about this topic, Information is NOT uncertainty!
Tributes to Claude Shannon, father of information theory, show the incredible impact of information theory on modern society.
![]()

Schneider Lab
origin: before 1996 March 9
updated:
2011 Jul 14
![]()