from Tom Schneider's Lab
organized by topic
-
Papers marked by:
 are available here
are available here
 are only available here.
are only available here.
 are only available
in the package of papers we NO LONGER send to people
or from your library.
are only available
in the package of papers we NO LONGER send to people
or from your library.
 are only available
from your library or online when a link is provided (you may need
a subscription).
are only available
from your library or online when a link is provided (you may need
a subscription).
 are from
my collaborators; I am not an author.
are from
my collaborators; I am not an author.
- My complete bibliography, containing the complete references for the papers mentioned below and older papers is given in BiBTeX format here.
- Current Bibliography in Medline Molecular Biology Subset. This does not include all papers, but does give many abstracts.
- All PostScript documents require a PostScript viewer or must be downloaded for printing.
- Books containing papers about our work
- Do you have problems printing these papers?

Theory of Molecular Machines
- Basic Math and Thermodynamics
 primer: Information Theory Primer
With an Appendix on Logarithms
primer: Information Theory Primer
With an Appendix on Logarithms
Read this one first to get a solid start on understanding the rest of our papers.- The Primer is also in Italian!. Unfortunately it is in an awful, primitive format. :-(
 A Glossary for Molecular Information Theory
A Glossary for Molecular Information Theory
 secondlaw: An Equation for the Second Law of
Thermodynamics
secondlaw: An Equation for the Second Law of
Thermodynamics
- Reviews
 trieste1996:
New Approaches in Mathematical Biology:
Information Theory and Molecular Machines
A brief 9 page introduction to molecular
information theory.
trieste1996:
New Approaches in Mathematical Biology:
Information Theory and Molecular Machines
A brief 9 page introduction to molecular
information theory.
 2000 Oct 13: full html, pdf and ps are now available
2000 Oct 13: full html, pdf and ps are now available
 T. D. Schneider,
Sequence Logos, Machine/Channel Capacity, Maxwell's Demon, and Molecular
Computers: a Review of the Theory of Molecular Machines
Nanotechnology,
5,
1-18,
1994
T. D. Schneider,
Sequence Logos, Machine/Channel Capacity, Maxwell's Demon, and Molecular
Computers: a Review of the Theory of Molecular Machines
Nanotechnology,
5,
1-18,
1994


 T. D. Schneider,
Consensus Sequence Zen,
Applied Bioinformatics,
1 (3),
111-119,
2002.
T. D. Schneider,
Consensus Sequence Zen,
Applied Bioinformatics,
1 (3),
111-119,
2002.
 as of 2002 Dec 5.
as of 2002 Dec 5.
 T. D. Schneider,
Some Lessons for Molecular Biology from Information Theory,
in
Entropy Measures, Maximum Entropy Principle
and Emerging Applications,
Springer-Verlag,
New York,
229-237.
T. D. Schneider,
Some Lessons for Molecular Biology from Information Theory,
in
Entropy Measures, Maximum Entropy Principle
and Emerging Applications,
Springer-Verlag,
New York,
229-237.
 as of 2003 April 4.
as of 2003 April 4.
 T. D. Schneider,
Twenty years of Delila and molecular information theory,
Biological Theory
1 (3),
250-260.
T. D. Schneider,
Twenty years of Delila and molecular information theory,
Biological Theory
1 (3),
250-260.
 as of 2007 Jan 12.
as of 2007 Jan 12.
 T. D. Schneider,
A Brief Review of Molecular Information Theory ,
Nano Communication Networks,
1: 173-180",
T. D. Schneider,
A Brief Review of Molecular Information Theory ,
Nano Communication Networks,
1: 173-180",
 as of 2011 Aug 11.
as of 2011 Aug 11.
- Books

Visualizing Biological Information by Clifford A. Pickover. @inproceedings{Rogan.Schneider1995, author = "P. K. Rogan and J. J. Salvo and R. M. Stephens and T. D. Schneider", title = "Visual Display of Sequence Conservation as an Aid to Taxonomic Classification Using {PCR} Amplification", editor = "Clifford A. Pickover", booktitle = "Visualizing Biological Information", publisher = "World Scientific", address = "Singapore", pages = "21-32", isbn = "ISBN 981-02-1427-8", year = "1995"}
Dictionary of Bioinformatics and Computational Biology, Hancock, John M. / Zvelebil, Marketa J. (eds.). (John Wiley & Sons, Inc., Hoboken, New Jersey, ISBN 0-471-43622-4, 2004.)
I (Thomas D. Schneider) am one of the contributors. The entries in the book correspond to some of those in A Glossary for Molecular Information Theory.
 as of 2004 July 30.
as of 2004 July 30.
Please note that there is an error in the web links published in the book!
- Theory of Molecular Machines Level 0:
Rsequence and Rfrequency
 T. D. Schneider,
G. D. Stormo,
L. Gold,
and A. Ehrenfeucht,
Information content of binding sites on nucleotide sequences,
J. Mol. Biol.,
188,
415-431,
1986.
T. D. Schneider,
G. D. Stormo,
L. Gold,
and A. Ehrenfeucht,
Information content of binding sites on nucleotide sequences,
J. Mol. Biol.,
188,
415-431,
1986.
 as of 2001 June 26
as of 2001 June 26
 T. D. Schneider,
G. J. Erickson and C. R. Smith (eds),
Information and entropy of patterns in genetic switches,
In: Maximum-Entropy and Bayesian Methods in Science and Engineering,
2,
147-154,
Kluwer Academic Publishers,
Dordrecht, The Netherlands,
1988.
T. D. Schneider,
G. J. Erickson and C. R. Smith (eds),
Information and entropy of patterns in genetic switches,
In: Maximum-Entropy and Bayesian Methods in Science and Engineering,
2,
147-154,
Kluwer Academic Publishers,
Dordrecht, The Netherlands,
1988.
 T. D. Schneider
and G. D. Stormo,
Excess Information at Bacteriophage T7 Genomic Promoters
Detected by a Random Cloning Technique,
Nucl. Acids Res.,
17,
659-674,
1989.
T. D. Schneider
and G. D. Stormo,
Excess Information at Bacteriophage T7 Genomic Promoters
Detected by a Random Cloning Technique,
Nucl. Acids Res.,
17,
659-674,
1989.
 N. D. Herman
and T. D. Schneider,
High Information Conservation Implies that at Least Three Proteins
Bind Independently to F Plasmid incD Repeats,
J. Bact.,
174,
3558-3560,
1992.
N. D. Herman
and T. D. Schneider,
High Information Conservation Implies that at Least Three Proteins
Bind Independently to F Plasmid incD Repeats,
J. Bact.,
174,
3558-3560,
1992.

 T. D. Schneider,
"Evolution of Biological Information",
Nucleic Acids Research,
28(14): 2794-2799, 2000
T. D. Schneider,
"Evolution of Biological Information",
Nucleic Acids Research,
28(14): 2794-2799, 2000
 as of 2000 July 4
as of 2000 July 4
Multiple Alignment
 T. D. Schneider
and D. Mastronarde,
Fast multiple alignment of ungapped DNA
sequences using information theory and a relaxation method,
Discrete Applied Mathematics,
71:
259-268,
1996
T. D. Schneider
and D. Mastronarde,
Fast multiple alignment of ungapped DNA
sequences using information theory and a relaxation method,
Discrete Applied Mathematics,
71:
259-268,
1996
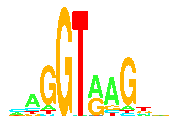
Sequence Logos  T. D. Schneider and R. M. Stephens,
Sequence
Logos: A New Way to Display Consensus Sequences
Nucl. Acids Res.,
18:
6097-6100,
1990
T. D. Schneider and R. M. Stephens,
Sequence
Logos: A New Way to Display Consensus Sequences
Nucl. Acids Res.,
18:
6097-6100,
1990
 M. C. Shaner
I. M. Blair,
and T. D. Schneider",
(T. N. Mudge and V. Milutinovic and L. Hunter, eds),
Sequence Logos: A Powerful, Yet Simple, Tool,
Proceedings of the Twenty-Sixth Annual Hawaii International
Conference on System Sciences, Volume 1: Architecture and Biotechnology
Computing",
813-821",
IEEE Computer Society Press",
Los Alamitos, CA",
1993
From the
``Alternative Approaches to Sequence Representation''
minitrack of the Biotechnology Computing Track,
Hawaii International Conference on System Sciences - 26,
Kauai, Hawaii - January 5-8, 1993
M. C. Shaner
I. M. Blair,
and T. D. Schneider",
(T. N. Mudge and V. Milutinovic and L. Hunter, eds),
Sequence Logos: A Powerful, Yet Simple, Tool,
Proceedings of the Twenty-Sixth Annual Hawaii International
Conference on System Sciences, Volume 1: Architecture and Biotechnology
Computing",
813-821",
IEEE Computer Society Press",
Los Alamitos, CA",
1993
From the
``Alternative Approaches to Sequence Representation''
minitrack of the Biotechnology Computing Track,
Hawaii International Conference on System Sciences - 26,
Kauai, Hawaii - January 5-8, 1993
 T. D. Schneider,
Reading of DNA Sequence Logos:
Prediction of Major Groove Binding
by Information Theory,
Meth. Enzym.,
274:
445-455,
1996.
T. D. Schneider,
Reading of DNA Sequence Logos:
Prediction of Major Groove Binding
by Information Theory,
Meth. Enzym.,
274:
445-455,
1996.
Advanced Logoology
 R. M. Stephens
and T. D. Schneider,
Features of spliceosome evolution and function
inferred from an analysis of the information at human splice sites,
J. Mol. Biol.,
228:
1124-1136,
1992
R. M. Stephens
and T. D. Schneider,
Features of spliceosome evolution and function
inferred from an analysis of the information at human splice sites,
J. Mol. Biol.,
228:
1124-1136,
1992
 P. P. Papp,
D. K. Chattoraj
and T. D. Schneider,
Information Analysis of Sequences that Bind
the Replication Initiator RepA,
J. Mol. Biol.,
233,
219-230,
1993.
PUBMED
P. P. Papp,
D. K. Chattoraj
and T. D. Schneider,
Information Analysis of Sequences that Bind
the Replication Initiator RepA,
J. Mol. Biol.,
233,
219-230,
1993.
PUBMED
 D. Barrick,
K. Villanueba,
J. Childs,
R. Kalil,
T. D. Schneider,
C. E. Lawrence,
L. Gold
and G. D. Stormo
Quantitative Analysis of Ribosome Binding Sites in
E. coli.,
Nucleic Acids Res.,
22,
1287-1295,
1994.
D. Barrick,
K. Villanueba,
J. Childs,
R. Kalil,
T. D. Schneider,
C. E. Lawrence,
L. Gold
and G. D. Stormo
Quantitative Analysis of Ribosome Binding Sites in
E. coli.,
Nucleic Acids Res.,
22,
1287-1295,
1994.
 M. B. Toledano,
I. Kullik,
F. Trinh,
P. T. Baird,
T. D. Schneider,
and G. Storz,
Redox-Dependent Shift of OxyR-DNA Contacts Along
An Extended {DNA} Binding Site:
A Mechanism for Differential Promoter Selection,
Cell,
78,
897-909,
1994.
M. B. Toledano,
I. Kullik,
F. Trinh,
P. T. Baird,
T. D. Schneider,
and G. Storz,
Redox-Dependent Shift of OxyR-DNA Contacts Along
An Extended {DNA} Binding Site:
A Mechanism for Differential Promoter Selection,
Cell,
78,
897-909,
1994.
 Replication control of plasmid P1
and its host chromosome: the
common ground,
D. K. Chattoraj and T. D. Schneider,
Prog. Nucl. Acid
Res. Mol. Biol., 57: 145-186,
chapter 5, Moldave (ed), 1997
Replication control of plasmid P1
and its host chromosome: the
common ground,
D. K. Chattoraj and T. D. Schneider,
Prog. Nucl. Acid
Res. Mol. Biol., 57: 145-186,
chapter 5, Moldave (ed), 1997
 R. K. Shultzaberger
and T. D. Schneider,
Using sequence logos and information analysis of Lrp DNA binding
sites to investigate discrepancies between natural selection and SELEX,
Nucleic Acids Res,
27 (3): 882-887, 1999.
R. K. Shultzaberger
and T. D. Schneider,
Using sequence logos and information analysis of Lrp DNA binding
sites to investigate discrepancies between natural selection and SELEX,
Nucleic Acids Res,
27 (3): 882-887, 1999.
 as of 1999 January 4
as of 1999 January 4
 N. Kannan,
T. D. Schneider
and S. Vishveshwara,
Logos for amino acid preferences in different backbone packing
density regions of protein structural classes,
Acta Crystallographica Section D,
56 (9), 1156-1165, 2000
N. Kannan,
T. D. Schneider
and S. Vishveshwara,
Logos for amino acid preferences in different backbone packing
density regions of protein structural classes,
Acta Crystallographica Section D,
56 (9), 1156-1165, 2000
 as of 2000 August 25
as of 2000 August 25

2001 December 1 issue of
Nucleic Acids Research
Copyright ® 2001
Oxford University Press. baseflip:
Strong Minor Groove Base Conservation in Sequence Logos
implies DNA Distortion or
Base Flipping during Replication and Transcription Initiation,
T. D. Schneider,
Nucl. Acid Res.,
29,
(23),
4881-4891,
2001.
baseflip:
Strong Minor Groove Base Conservation in Sequence Logos
implies DNA Distortion or
Base Flipping during Replication and Transcription Initiation,
T. D. Schneider,
Nucl. Acid Res.,
29,
(23),
4881-4891,
2001.
 as of 2001 Nov 28
as of 2001 Nov 28
 repan3:
The P1 Phage Replication Protein RepA Contacts an Otherwise Inaccessible
Thymine N3 Proton by DNA Distortion or Base Flipping,
I. G. Lyakhov,
P. N. Hengen,
D. Rubens
and T. D. Schneider
Nucl. Acid Res.,
29,
(23),
4892-4900
2001.
repan3:
The P1 Phage Replication Protein RepA Contacts an Otherwise Inaccessible
Thymine N3 Proton by DNA Distortion or Base Flipping,
I. G. Lyakhov,
P. N. Hengen,
D. Rubens
and T. D. Schneider
Nucl. Acid Res.,
29,
(23),
4892-4900
2001.
 as of 2001 Nov 28
as of 2001 Nov 28

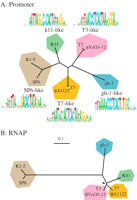 Z. Chen and
T. D. Schneider,
Information theory based T7-like promoter models: classification of
bacteriophages and differential evolution of promoters and their
polymerases,
Nucleic Acids Res,
33,
6172-6187,
2005,
Z. Chen and
T. D. Schneider,
Information theory based T7-like promoter models: classification of
bacteriophages and differential evolution of promoters and their
polymerases,
Nucleic Acids Res,
33,
6172-6187,
2005,
 as of 2005 Dec 14.
as of 2005 Dec 14.
 E. Bindewald,
T. D. Schneider
and B. A. Shapiro,
CorreLogo: An online server for 3D sequence logos of RNA and
DNA alignments,
Nucleic Acids Res.
34,
w405-w411,
2006.
E. Bindewald,
T. D. Schneider
and B. A. Shapiro,
CorreLogo: An online server for 3D sequence logos of RNA and
DNA alignments,
Nucleic Acids Res.
34,
w405-w411,
2006.
 as of 2006 Jul 19.
as of 2006 Jul 19.

Individual Information and sequence walkers 
 ri:
Schneider T. D.
Information content of individual genetic sequences.
J. Theor. Biol. 189(4): 427-441, 1997.
ri:
Schneider T. D.
Information content of individual genetic sequences.
J. Theor. Biol. 189(4): 427-441, 1997.
 as of 1997 Dec 23.
as of 1997 Dec 23.
 was not available prior to 1998 April 26.
was not available prior to 1998 April 26.
 walker:
Schneider T. D.
Sequence walkers: a graphical method to display how binding proteins
interact with DNA or RNA sequences,
Nucl. Acids Res. 25: 4408-4415, 1997.
walker:
Schneider T. D.
Sequence walkers: a graphical method to display how binding proteins
interact with DNA or RNA sequences,
Nucl. Acids Res. 25: 4408-4415, 1997.
 as of 1997 Nov 24.
as of 1997 Nov 24.
 was not available prior to 1998 April 26.
was not available prior to 1998 April 26.
 fisinfo:
P. N. Hengen, S. L.
Bartram, L. E. Stewart, T. D. Schneider,
Information Analysis of Fis
Binding Sites, Nucl. Acids Res. 25(24): 4994-5002, 1997.
fisinfo:
P. N. Hengen, S. L.
Bartram, L. E. Stewart, T. D. Schneider,
Information Analysis of Fis
Binding Sites, Nucl. Acids Res. 25(24): 4994-5002, 1997.
 as of 1998 Jan 8.
as of 1998 Jan 8.
 was not available prior to 1998 April 26.
was not available prior to 1998 April 26.
 oxyrfur:
M. Zheng, B. Doan, T. D. Schneider, and G. Storz,
OxyR and SoxRS Regulation of fur,
J. Bact.,
181 (15): 4639-4643.
oxyrfur:
M. Zheng, B. Doan, T. D. Schneider, and G. Storz,
OxyR and SoxRS Regulation of fur,
J. Bact.,
181 (15): 4639-4643.
 as of 1999 July 26
as of 1999 July 26
 lrp:
Using sequence logos and information analysis of Lrp DNA binding
sites to investigate discrepancies between natural selection and SELEX,
Nucl. Acids Res. 27(3): 882-887, 1999.
lrp:
Using sequence logos and information analysis of Lrp DNA binding
sites to investigate discrepancies between natural selection and SELEX,
Nucl. Acids Res. 27(3): 882-887, 1999.
 as of 2001 Jun 6.
as of 2001 Jun 6.
 ridebate:
T. D. Schneider,
Measuring
Molecular Information,
Journal of Theoretical Biology
201:
87-92,
1999.
ridebate:
T. D. Schneider,
Measuring
Molecular Information,
Journal of Theoretical Biology
201:
87-92,
1999.
 as of 1999 November 3
as of 1999 November 3
 SoxS:
"Interdependence of the Position and Orientation of
SoxS Binding Sites in the
Transcriptional Activation of the Class I Subset of
Escherichia coli
Superoxide-Inducible Promoters",
T. I. Wood,
K. L. Griffith,
W. P. Fawcett,
K.-W. Jair,
T. D. Schneider,
and R. E. Wolf,
Molec. Microb.,
34(3): 414-430.
SoxS:
"Interdependence of the Position and Orientation of
SoxS Binding Sites in the
Transcriptional Activation of the Class I Subset of
Escherichia coli
Superoxide-Inducible Promoters",
T. I. Wood,
K. L. Griffith,
W. P. Fawcett,
K.-W. Jair,
T. D. Schneider,
and R. E. Wolf,
Molec. Microb.,
34(3): 414-430.
 as of 1999 December 20.
Supplementary
information is available.
as of 1999 December 20.
Supplementary
information is available.
 zheng.storz2001.
Journal of Bacteriology, August 2001, p.
4571-4579, Vol. 183, No. 15,
"Computation-Directed
Identification of OxyR
DNA Binding Sites in
Escherichia coli"
Ming Zheng, Xunde Wang,
Bernard Doan, Karen A. Lewis, Thomas D. Schneider, and Gisela
Storz.
zheng.storz2001.
Journal of Bacteriology, August 2001, p.
4571-4579, Vol. 183, No. 15,
"Computation-Directed
Identification of OxyR
DNA Binding Sites in
Escherichia coli"
Ming Zheng, Xunde Wang,
Bernard Doan, Karen A. Lewis, Thomas D. Schneider, and Gisela
Storz.
 R. K. Shultzaberger,
R. E. Bucheimer,
K. E. Rudd
and T. D. Schneider,
Anatomy of Escherichia coli Ribosome Binding Sites
J. Mol. Biol., 313: 215-228, 2001.
flexrbs
R. K. Shultzaberger,
R. E. Bucheimer,
K. E. Rudd
and T. D. Schneider,
Anatomy of Escherichia coli Ribosome Binding Sites
J. Mol. Biol., 313: 215-228, 2001.
flexrbs
 as of 2001 September 25, published
2001 October 16.
as of 2001 September 25, published
2001 October 16.

 colonsplice:
P. K. Rogan and T. D. Schneider,
Using Information Content and Base
Frequencies to Distinguish Mutations from Genetic Polymorphisms in Splice
Junction Recognition Sites.
Human Mutation, 6: 74-76, 1995
colonsplice:
P. K. Rogan and T. D. Schneider,
Using Information Content and Base
Frequencies to Distinguish Mutations from Genetic Polymorphisms in Splice
Junction Recognition Sites.
Human Mutation, 6: 74-76, 1995
 rfs:
P. K. Rogan, B. M. Faux and T. D. Schneider,
Information analysis of human splice site mutations
Human Mutation
12(3):
153-171 (1998).
rfs:
P. K. Rogan, B. M. Faux and T. D. Schneider,
Information analysis of human splice site mutations
Human Mutation
12(3):
153-171 (1998).
 as of 1998 May 15.
as of 1998 May 15.
 abcr:
Rando Allikmets, Wyeth W. Wasserman, Amy
Hutchinson, Philip Smallwood, Jeremy Nathans,
Peter K. Rogan, Thomas D. Schneider, Michael
Dean,
Organization of the ABCR gene: analysis of promoter and splice junction
sequences,
Gene (215)1: 111-122 , 1998.
abcr:
Rando Allikmets, Wyeth W. Wasserman, Amy
Hutchinson, Philip Smallwood, Jeremy Nathans,
Peter K. Rogan, Thomas D. Schneider, Michael
Dean,
Organization of the ABCR gene: analysis of promoter and splice junction
sequences,
Gene (215)1: 111-122 , 1998.
 as of 1998 October 16.
as of 1998 October 16.
 S. G. Kahn, H. L. Levy, R. Legerski, E. Quackenbush, J. T. Reardon, S. Emmert,
A. Sancar, L. Li, T. D. Schneider, J. E. Cleaver, and K. H. Kraemer.
Xeroderma Pigmentosum Group C splice mutation associated with
mutism and hypoglycinemia - A new syndrome?
Journal of Investigative Dermatology, 111:791-796, 1998.
S. G. Kahn, H. L. Levy, R. Legerski, E. Quackenbush, J. T. Reardon, S. Emmert,
A. Sancar, L. Li, T. D. Schneider, J. E. Cleaver, and K. H. Kraemer.
Xeroderma Pigmentosum Group C splice mutation associated with
mutism and hypoglycinemia - A new syndrome?
Journal of Investigative Dermatology, 111:791-796, 1998.
 as of 1998 November 2
as of 1998 November 2
 Splice Site Mutations in Atherosclerosis Candidate Genes:
Relating Individual Information to Phenotype.
Yskert von Kodolitsch, Reed E. Pyeritz, and Peter K. Rogan,
Circulation
100: 693-699, 1999,
Splice Site Mutations in Atherosclerosis Candidate Genes:
Relating Individual Information to Phenotype.
Yskert von Kodolitsch, Reed E. Pyeritz, and Peter K. Rogan,
Circulation
100: 693-699, 1999,
 in press as of 1999 May 27;
published as of 1999 August 16
in press as of 1999 May 27;
published as of 1999 August 16
 Exon Skipping in IVD RNA Processing in Isovaleric Acidemia Caused
by Point Mutations in the Coding Region of the IVD Gene,
Jerry Vockley, Peter K. Rogan, Bambi D. Anderson, Jan Willard, Ratnam
S. Seelan, David I. Smith, and Wanguo Liu,
American
Journal of Human Genetics
66:356-367, 2000
Exon Skipping in IVD RNA Processing in Isovaleric Acidemia Caused
by Point Mutations in the Coding Region of the IVD Gene,
Jerry Vockley, Peter K. Rogan, Bambi D. Anderson, Jan Willard, Ratnam
S. Seelan, David I. Smith, and Wanguo Liu,
American
Journal of Human Genetics
66:356-367, 2000
 as of 2000 March 2
as of 2000 March 2
 Redundant Designations of BRCA1 Intron 11 Splicing Mutation,
Stan R. Svojanovsky, Thomas D. Schneider, and Peter K. Rogan.
Human
Mutation,
16: 264 (2000)
Redundant Designations of BRCA1 Intron 11 Splicing Mutation,
Stan R. Svojanovsky, Thomas D. Schneider, and Peter K. Rogan.
Human
Mutation,
16: 264 (2000)
 as of 2000 Sep 8
as of 2000 Sep 8
 I. Arnould et al,
Identifying and characterizing a five-gene cluster
of ATP-binding cassette transporters mapping to human
chromosome 17q24: a new subgroup within the ABCA subfamily,
GeneScreen,
1:
157-164, 2001.
I. Arnould et al,
Identifying and characterizing a five-gene cluster
of ATP-binding cassette transporters mapping to human
chromosome 17q24: a new subgroup within the ABCA subfamily,
GeneScreen,
1:
157-164, 2001.
 as of 2001 November 19.
as of 2001 November 19.
 Khan.Kraemer2002:
Khan SG, Muniz-Medina V, Shahlavi T, Baker CC, Inui
H, Ueda T, Emmert S, Schneider TD, Kraemer KH.
The human XPC DNA repair gene: arrangement, splice site information
content and influence of a single nucleotide polymorphism in a splice
acceptor site on alternative splicing and function.
Nucleic Acids Res. 2002 Aug 15;30(16):3624-31. Pubmed PMID:
12177305.
Khan.Kraemer2002:
Khan SG, Muniz-Medina V, Shahlavi T, Baker CC, Inui
H, Ueda T, Emmert S, Schneider TD, Kraemer KH.
The human XPC DNA repair gene: arrangement, splice site information
content and influence of a single nucleotide polymorphism in a splice
acceptor site on alternative splicing and function.
Nucleic Acids Res. 2002 Aug 15;30(16):3624-31. Pubmed PMID:
12177305.  as of
2002 August 28.
as of
2002 August 28.  Rogan PK, Svojanovsky S, Leeder JS.
Information theory-based analysis of CYP2C19,
CYP2D6 and CYP3A5 splicing mutations,
Pharmacogenetics 2003 13(4): 207-18.
Rogan PK, Svojanovsky S, Leeder JS.
Information theory-based analysis of CYP2C19,
CYP2D6 and CYP3A5 splicing mutations,
Pharmacogenetics 2003 13(4): 207-18.
 as of 2003 April 7.
as of 2003 April 7.

 P. N. Hengen,
I. G. Lyakhov,
L. E. Stewart
and T. D. Schneider,
Molecular Flip-Flops Formed
by Overlapping Fis Sites,
Nucleic Acids Res.,
31:
6663-6673,
2003
P. N. Hengen,
I. G. Lyakhov,
L. E. Stewart
and T. D. Schneider,
Molecular Flip-Flops Formed
by Overlapping Fis Sites,
Nucleic Acids Res.,
31:
6663-6673,
2003
 as of 2003 October 15.
as of 2003 October 15.
 Two Essential Splice Lariat Branchpoint Sequences In One
Intron in a Xeroderma Pigmentosum DNA Repair Gene:
Mutations Result in
Reduced XPC mRNA Levels that Correlate with Cancer Risk,
Khan SG, Metin A, Gozukara E, Inui H, Shahlavi T, Muniz-Medina V,
Baker CC, Ueda T, Aiken JR, Schneider TD, Kraemer KH,
Hum Mol Genet. 2004 Feb 1; 13(3): 343-352.
Two Essential Splice Lariat Branchpoint Sequences In One
Intron in a Xeroderma Pigmentosum DNA Repair Gene:
Mutations Result in
Reduced XPC mRNA Levels that Correlate with Cancer Risk,
Khan SG, Metin A, Gozukara E, Inui H, Shahlavi T, Muniz-Medina V,
Baker CC, Ueda T, Aiken JR, Schneider TD, Kraemer KH,
Hum Mol Genet. 2004 Feb 1; 13(3): 343-352.
 as of 2004 January 18.
as of 2004 January 18.
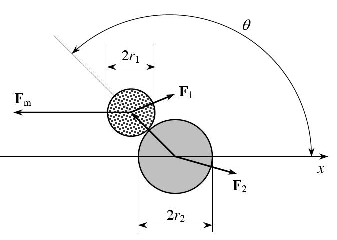
Theory of Molecular Machines
- Theory of Molecular Machines Level 1:
 T. D. Schneider,
Theory of Molecular Machines.
I. Channel Capacity of Molecular Machines
J. Theor. Biol.,
148:
83-123,
1991
T. D. Schneider,
Theory of Molecular Machines.
I. Channel Capacity of Molecular Machines
J. Theor. Biol.,
148:
83-123,
1991
- Theory of Molecular Machines Level 2:
 T. D. Schneider,
Theory of Molecular Machines.
II. Energy Dissipation from Molecular Machines
J. Theor. Biol.,
148:
125-137,
1991
T. D. Schneider,
Theory of Molecular Machines.
II. Energy Dissipation from Molecular Machines
J. Theor. Biol.,
148:
125-137,
1991

 T. D. Schneider,
Claude Shannon: Biologist,
25,
(1) (January/February),
30-33,
2006,
IEEE Engineering in Medicine and Biology Magazine
T. D. Schneider,
Claude Shannon: Biologist,
25,
(1) (January/February),
30-33,
2006,
IEEE Engineering in Medicine and Biology Magazine
 as of 2005 Dec 27.
as of 2005 Dec 27.
 T. D. Schneider,
70% efficiency of bistate molecular machines explained by information
theory, high dimensional geometry and evolutionary convergence
T. D. Schneider,
70% efficiency of bistate molecular machines explained by information
theory, high dimensional geometry and evolutionary convergence
 as of 2010 Apr 30.
as of 2010 Apr 30.
 T. D. Schneider,
A Brief Review of Molecular Information Theory
T. D. Schneider,
A Brief Review of Molecular Information Theory
 as of 2011 Jul 15.
as of 2011 Jul 15.
 Thomas D. Schneider and Vishnu Jejjala
Restriction enzymes use a 24 dimensional coding space to recognize 6
base long DNA sequences
Thomas D. Schneider and Vishnu Jejjala
Restriction enzymes use a 24 dimensional coding space to recognize 6
base long DNA sequences
 as of 2020 Mar 16.
as of 2020 Mar 16.
|
|

|
 The bottle,
a story published in
Nature 406: 351, 27 July 2000
The bottle,
a story published in
Nature 406: 351, 27 July 2000
 as of 2001 January 24
as of 2001 January 24
 I. G. Lyakhov,
T. D. Schneider,
G. A. Lyakhov
and N. V. Suyazov,
Orientational Ordering of Protein Micro-
and Nanoparticles in a Nonuniform Magnetic Field,
Physics of Wave Phenomena
13:1-14, 2005.
I. G. Lyakhov,
T. D. Schneider,
G. A. Lyakhov
and N. V. Suyazov,
Orientational Ordering of Protein Micro-
and Nanoparticles in a Nonuniform Magnetic Field,
Physics of Wave Phenomena
13:1-14, 2005.
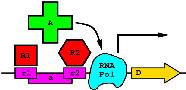

 Patent
on how to build a
molecular computer:
Molecular Computing Elements: Gates and Flip-Flops
by TD Schneider and PN Hengen
Patent
on how to build a
molecular computer:
Molecular Computing Elements: Gates and Flip-Flops
by TD Schneider and PN Hengen
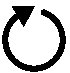
 Patent pending on
a molecular motor:
Molecular Rotation Engine
by
Thomas D. Schneider and Ilya G. Lyakhov
Patent pending on
a molecular motor:
Molecular Rotation Engine
by
Thomas D. Schneider and Ilya G. Lyakhov
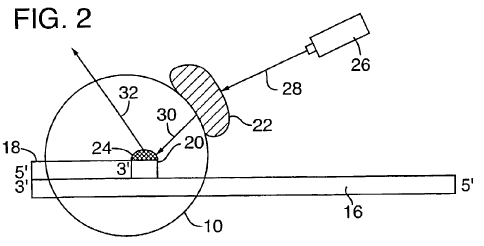
 Patent pending on
a method of
molecular sequencing:
High Speed Parallel Nucleic Acid Sequencing
by Thomas D. Schneider and Denise Rubens.
Patent pending on
a method of
molecular sequencing:
High Speed Parallel Nucleic Acid Sequencing
by Thomas D. Schneider and Denise Rubens.
![]()

Schneider Lab
origin: 1996 Feb 15
updated: version = 2.26 of webpapers.html 2020 Mar 16
![]()