|
Summary in layperson's terms
Cell phones and molecules face the same task: figuring out a signal in
the presence of overwhelming noise.
This is the story of why your cell phone gives you such a clear signal
that you don't realize that it is fighting to figure out the desired
signal from noisy electrical interference. It is also the story of
how a tiny protein molecule can find exactly one sequence on DNA, such
as GAATTC, despite noisy interference in that task by being bombarded
by the surrounding water molecules and ions. Mathematically they are
the same story.
When you talk on your cell phone, a signal comes from the other person
into the electronics. The signal initially is clean voltage pulses,
e.g. exactly 1 and 0 volts. But by the time it gets to your
phone it is degraded by noise. Heat in the wires, called thermal
noise, kicks electrons to higher or lower energy. A series of kicks
happen before you get each pulse. So sometimes you would get 1.1
volts, another time 0.98 and so on. There is a law in statistics
called the "central limit theorem" that says that whenever a large
number of random values are added together, the distribution will
follow a "bell shaped curve" (also known more precisely as a Normal or
Gaussian distribution). So if you made a histogram of the voltages of
the pulses, you would get a Gaussian distribution.
If you were to receive a long message and measure the voltage for
every 0 pulse sent, a graph of the pulse voltages you observe would be
Gaussian along a line, centered at zero volts. Likewise, the 1 pulses
would also be Gaussian, but centered at 1 volt.
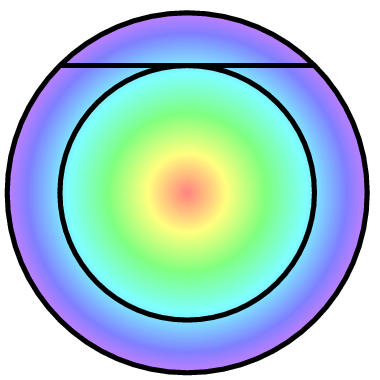
Now consider the first two pulses of a message. Each pulse and its
noise is independent from the other pulse. Geometrically two
independent values can be plotted at right angles. It turns out that
the distribution of two Gaussian distributions combined is circularly
symmetric. (You can work out why that is as homework or you can read
at
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1538977/
or
https://alum.mit.edu/www/toms/papers/shannonbiologist/
- see the
box, "Representing a message as a hypersphere.") That is, it's a
fuzzy circle. With three pulses, one gets a spherical distribution.
With four pulses ... it's a 4 dimensional "hypersphere". Don't
panic! Just think in three dimensions and do mathematics to figure
out details. It turns out that as one keeps adding pulses, the
distribution becomes more and more like a ping pong ball with
everything on the surface (See
https://alum.mit.edu/www/toms/papers/ccmm/
for the mathematical
details.)
So if I send you a message as a series of pulses, that can be
represented as a point in a high dimensional space. However, noise
interferes with every pulse on every dimensional axis, so you get a
point on the surface of a hypersphere around the original message
point. Claude Shannon's brilliant realization was that you could
figure out the original noise-free message by picking the center of
the hypersphere. That removes the noise and it explains why your cell
phone communications are so clear, despite the noise! The process of
finding the nearest hypersphere center is called 'decoding'.
However, this scheme will only work if the messages are far enough
apart in the high dimensional space that they don't intersect. When
you get a message with noise (a point on the surface of a hypersphere
around the original message point) you have to pick the nearest
possible message, and that won't always work if the spheres overlap.
So the problem becomes one of how to pack spheres together in a high
dimensional space so that the communications works with as few errors
as possible. Obviously this has been solved and it is the basis of
our communications systems.
So how best to pack spheres together? In two dimensions one could put
coins down in a square array, but we all know that they pack together
better if one uses a hexagonal array
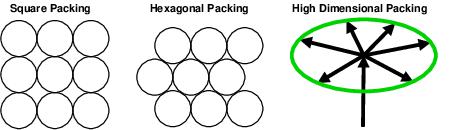 (See the first figure in our
paper at
https://doi.org/10.1371/journal.pone.0222419).
A pile of oranges in a food
store is the best way to pack spheres in three dimensions.
(See the first figure in our
paper at
https://doi.org/10.1371/journal.pone.0222419).
A pile of oranges in a food
store is the best way to pack spheres in three dimensions.
For our paper, we are interested in understanding how biological
molecules work. It turns out that they use exactly the same
mathematics as Shannon developed for communications, information and
coding theory.
The protein EcoRI binds to DNA at the sequence GAATTC. To describe
this using information theory, just think about the first base, 'G'.
That is one of four possibilities. Now simplify the problem to just
head's and tails of a coin. If I have a coin on a table with heads or
tails facing up but you don't see it, you would have to ask me one
question "is it heads" and I would answer yes or no. That tells you
one bit of information. In the case of DNA there are four
possibilities, A, C, G and T so you could ask two questions: "Is it in
the set G or A?" and "Is it in the set G or T?" My two answers tell
you 2 bits of information.
We can do this for all 6 positions in the binding site GAATTC. Then
the total number of bits is 6 (positions per site) x 2 (bits per
position) = 12 (bits per site) since bits can be added, a property of
information that Shannon initially required of the mathematics.
An invading virus will inject its DNA into a bacterium and take over
the cell. Bacteria have developed a defense that "restricts" the
growth of the virus. These are the restriction enzymes. After the
virus has injected its DNA into a bacterium, EcoRI molecules
inside the cell will bind
to the DNA and cut it at GAATTC. This destroys the virus and so
protects the bacterium.
EcoRI essentially does not cut at any other sequence, and so it is a
useful reagent for genetic engineering that we use for cutting up DNA.
For EcoRI to bind to GAATTC, it needs a set of specific contacts to
the DNA. It's like a key in a lock. The lock has a number of
two-part pins that move up and down based on the key. If it's the
right key, the pins move so that the break between the pins are all at
the shear line and the lock can open. If two pins were to move
together, then the lock would be less secure because it would be
easier to pick. So the pins should move independently. But that
means that the state of the lock can be described by a set of
independent numbers and therefore a lock is a high dimensional device.
So how many 'pins' does EcoRI have? That is, how many dimensions does
EcoRI work in? That's what our paper answers.
I got the an equation for the dimensionality in 1994.
When I showed the equation to Vishnu Jejjala, who was a high
school student working with me at that time,
he said several smart things.
First, he noted that
the equation is a lower bound on the dimensionality.
Unfortunately, having
a lower bound is disappointing because the
higher the dimension, the sharper the spheres are (more like a ping
pong ball instead of a fuzzy ball of yarn). So in biology we would
expect a very high dimensionality and a lower bound is useless.
But then Vishnu suggested that there might also be an
upper bound equation and that the two equations might converge to give
a single answer to the what the dimensionality is.
18 years later (!) I was reading
a paper by Jaynes. He knew that
muscle is 70% efficient but the only tool he had to think about that
was the Carnot efficiency. This is the efficiency of a heat engine
as in a car. It uses a high temperature (burning fuel) and a low
temperature (the outside air) to drive the engine. Carnot's equation
does not apply to biological systems because those work at one
temperature.
For example, by holding onto the negative charge of the DNA backbone,
EcoRI can glide along DNA like a train on spiral tracks. Before EcoRI
is bound to DNA, it is somewhere along the DNA. It quickly goes to
equilibrium with the surrounding water. That is, it is at the same
temperature as the water. Because it gets kicked around by thermal
noise, EcoRI moves along the DNA by random Brownian motion and
eventually when it encounters a GAATTC sequence, it forms contacts and
binds there. For it to stick, energy must be dissipated out into the
surrounding water. The amount of energy can be measured using a
microcalorimeter. To get some idea about how fast the energy leaves
the EcoRI/DNA complex, I looked into the speed of sound in sea water.
It turns out that it takes about a picosecond for sound to cross the
diameter of DNA. So in a few picoseconds the heat will go away and
the EcoRI will be bound to the DNA at the temperature of the
surroundings. The temperature before binding (Th) is the same as the
temperature after binding (Tc). As a result the Carnot efficiency
((Th - Tc)/Th) is zero, so that's not the right way to measure the
efficiency.
It turns out that there is an equation developed
from Shannon's work (the channel capacity equation)
by Pierce and Cutler
and published in 1959 that can be used when the temperature is
constant - an isothermal efficiency. Pierce wrote the best
introduction to information theory (Symbols Signals And Noise
https://archive.org/details/symbolssignalsan002575mbp).
Cutler was my
father's boss at Bell Labs. (I met him once when I was perhaps 5 - he
was as tall as a two story building and had white hair.) I used that
efficiency equation and hypersphere packing to explain why many
molecules, including EcoRI and the light sensitive protein in your eye
rhodopsin, are 70% efficient (see
https://alum.mit.edu/www/toms/papers/emmgeo/).
So when I saw the paper in which Jaynes was trying to use the Carnot
equation for muscle, I knew he was doomed. But I eagerly read the
paper to see how he would handle it and he did indeed fail because he
didn't have the Pierce / Cutler equation for isothermal efficiency.
But Jaynes was not stupid. He wrote so clearly that he lucidly
explained an equation for noise that I had already for 20 years. His
explanation led me to realize that the noise equation could be
rearranged to determine an upper bound for the dimensionality. Vishnu
was right, there really is an equation for an upper bound!
A few manipulations made both the lower and upper bounds look really
similar - a rather mysterious result - and when one asks how they
would evolve to get to 70% efficiency ... the upper and lower bounds
converge to a spectacularly simple answer: twice the information
measured in bits. So the 12 bit GAATTC that EcoRI binds to means it
works in 24 dimensions.
Again, Vishnu was right - the bounds converge. Essentially he was my
mentor all those years.
Since Shannon developed his theory of packing of high dimensional
spheres in 1949 people have been busy figuring out the best ways to
pack spheres in various dimensions. It turns out that the very best
way to pack spheres is called the Leech lattice, and it is in 24
dimensions.
In other words, we believe that EcoRI probably uses a Leech lattice to
find its binding sites precisely. This implies that we might be able
to build the noise decoder in cellphones using a single molecule.
|