We aligned the sequences of 149 E. coli and coliphage ribosome binding
sites by their initiation codons because the process of initiation requires
that the fmet-tRNA
![]() bind there. Since ribosomes search mRNA, we used the
composition of the transcript library (Stormo et al., 1982a)
to calculate Hg:
A=29526, C=25853, G=27800, T=28951 for which Hg=1.99817 bits/base. The
frequencies of bases at each position of the sites were used to find the
information content,
Rsequence(L),
as a function of position
(equations 2, 3 and a.8).
Fig. 2
shows that the largest peak is for the initiation codon.
The second largest peak represents the "Shine and Dalgarno" sequence (Shine
and Dalgarno, 1974). There are at least five other distinct peaks.
bind there. Since ribosomes search mRNA, we used the
composition of the transcript library (Stormo et al., 1982a)
to calculate Hg:
A=29526, C=25853, G=27800, T=28951 for which Hg=1.99817 bits/base. The
frequencies of bases at each position of the sites were used to find the
information content,
Rsequence(L),
as a function of position
(equations 2, 3 and a.8).
Fig. 2
shows that the largest peak is for the initiation codon.
The second largest peak represents the "Shine and Dalgarno" sequence (Shine
and Dalgarno, 1974). There are at least five other distinct peaks.
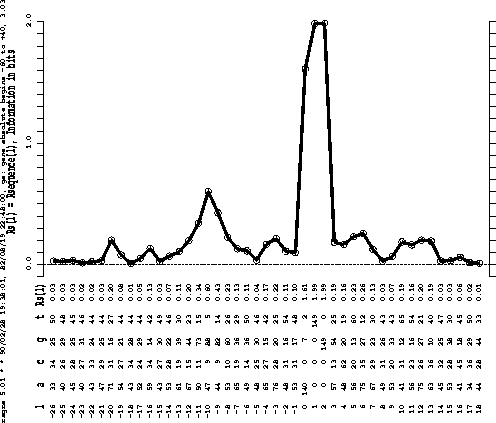
Position 0 is the first base of the initiation codon. |
Rsequence, the total information content of the site, is found by adding together the individual information contents from each position (equation 6). Previous statistical analyses showed a range of -21 to +13 (zero is the first base of the initiation codon), which corresponds well to the regions of RNA protected by ribosomes from ribonucleases (Gold et al., 1981). This range was extended by 5 bases on both sides. For this range, we calculate an Rsequence of 11.0 bits per site. Alignment by the Shine and Dalgarno sequence gives less than 8.3 bits (data not shown), which suggests that this is not a good alignment.
A good estimate for the size of the E. coli genome is
![]() basepairs
(Bachmann and Low, 1980). In determining
Rfrequency, we
assume that almost
all of the genome is transcribed into messages
and that for the most part only one strand is transcribed.
The number of potential ribosome binding sites is therefore
basepairs
(Bachmann and Low, 1980). In determining
Rfrequency, we
assume that almost
all of the genome is transcribed into messages
and that for the most part only one strand is transcribed.
The number of potential ribosome binding sites is therefore
![]() .
Based on the coding capacity versus
DNA insert size of 24 plasmids
selected at random from the Clark-Carbon bank (P. Bloch, personal
communication; F.C. Neidhardt et al., 1983),
and a genome size of
.
Based on the coding capacity versus
DNA insert size of 24 plasmids
selected at random from the Clark-Carbon bank (P. Bloch, personal
communication; F.C. Neidhardt et al., 1983),
and a genome size of
![]() bp, we estimate the number of proteins encoded by E. coli,
and therefore the
number of ribosome binding sites, to be 2574.
Equation (9) therefore gives an
Rfrequency of
10.6 bits per site. The data for all analyses are gathered in
Table 1.
bp, we estimate the number of proteins encoded by E. coli,
and therefore the
number of ribosome binding sites, to be 2574.
Equation (9) therefore gives an
Rfrequency of
10.6 bits per site. The data for all analyses are gathered in
Table 1.