

(citation will go here) Franco.Schneider2022 Kevin S. Franco, Zhe Sun, Yixiong Chen, Cedric Cagliero, Yuhong Zuo, Yan Ning Zhou, Mikhail Kashlev, Ding Jun Jin and Thomas D. Schneider
In E. coli, one RNA polymerase (RNAP) transcribes all RNA species, and different regulons are transcribed by employing different sigma (σ) factors. RNAP containing σ38 (σS) activates genes responding to stress conditions such as stationary phase. The structure of σ38 promoters has been controversial for more than two decades. To construct a model of σ38 promoters using information theory, we aligned proven transcriptional start sites to maximize the sequence information, in bits, and identified a -10 element similar to σ70 promoters. We could not align any -35 sequence logo; instead we found two patterns upstream of the -35 region. These patterns have dyad symmetry sequences and correspond to the location of UP elements in ribosomal RNA (rRNA) promoters. Additionally the UP element dyad symmetry suggests that the two polymerase α subunits, which bind to the UPs, should have two-fold dyad axis of symmetry on the polymerase and this is indeed observed in an X-ray crystal structure. Curiously the αCTDs should compete for overlapping UP elements. In vitro experiments confirm that σ38 recognizes the rrnB P1 promoter, requires a -10, UP elements and no -35. This clarifies the long-standing paradox of how σ38 promoters differ from those of σ70.
Proposed cover figure:
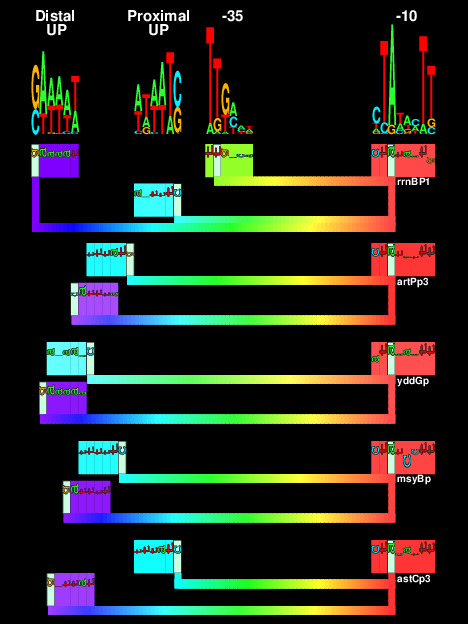
|
Proposed graphical abstract:
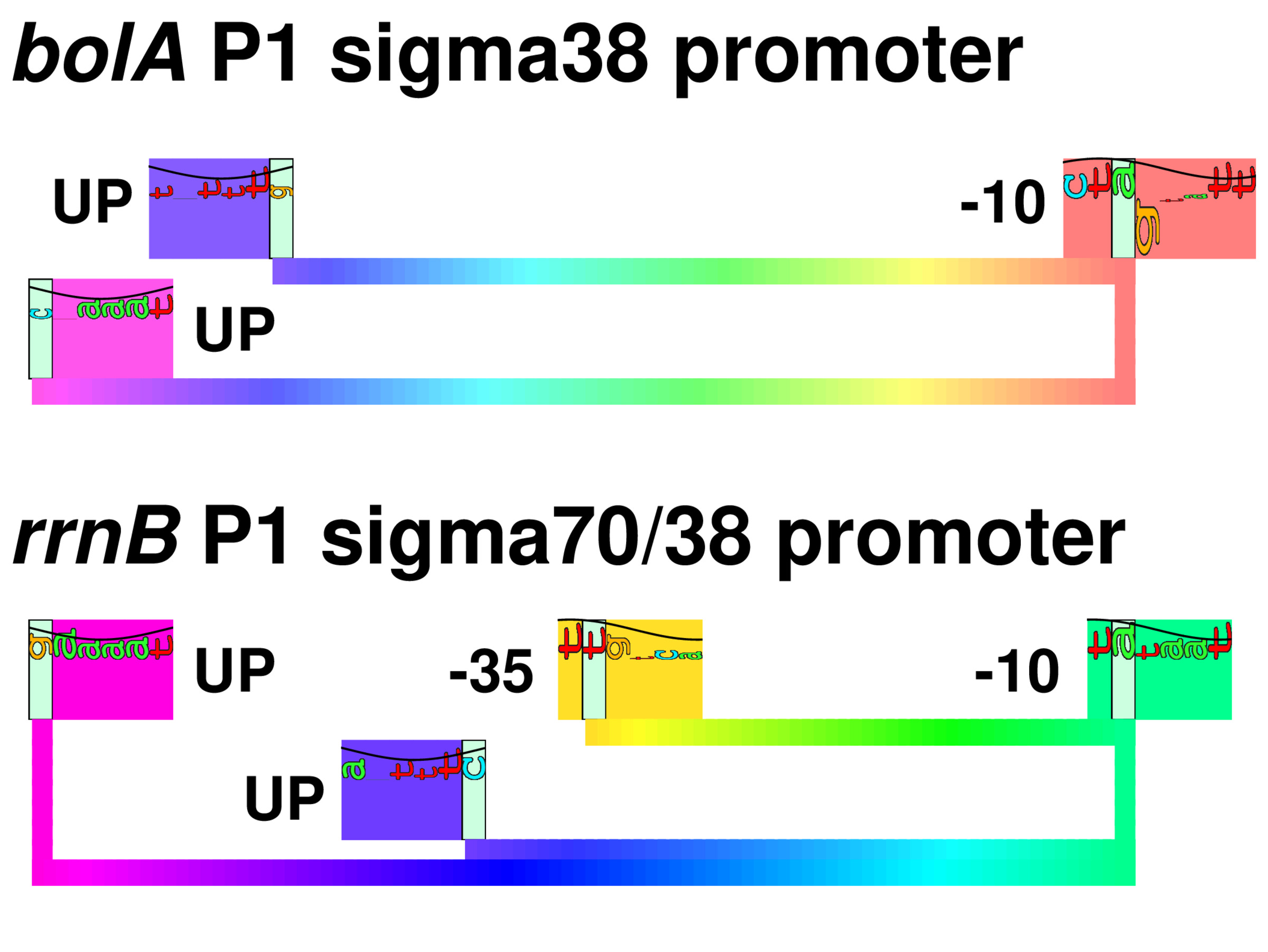
|
Permanent link https://alum.mit.edu/www/toms/papers/sigma38 points to the current web location.
![]()

Schneider Lab
origin: 2020 Feb 05
updated:
version = 1.03 of sigma38.html 2022 Jan 21
![]()