|
and the Theory of Molecular Machines |
|
Bits measure biological conservation! |
|
|
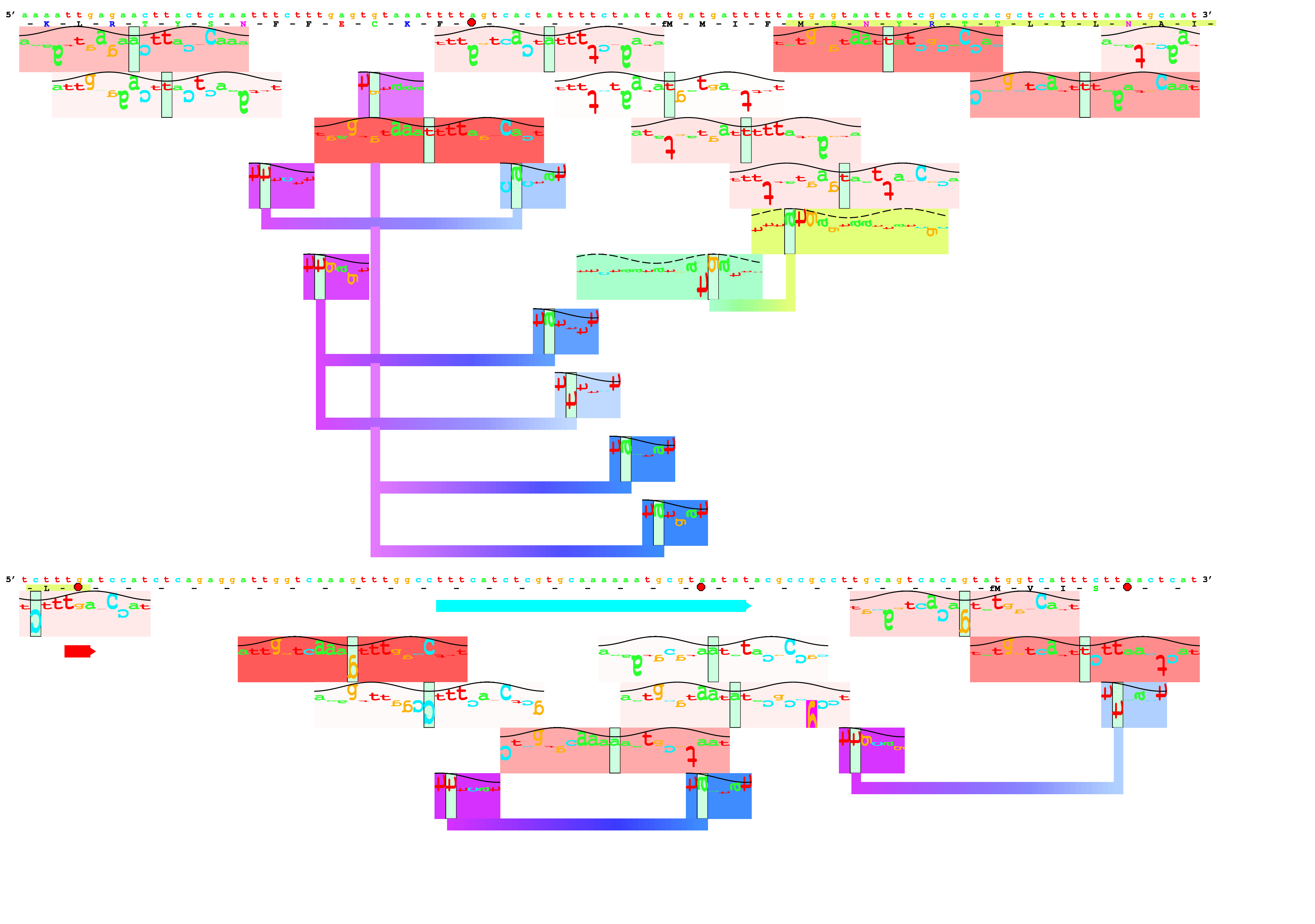
Brief overview: Biological Information Theory (BIT) is the application of Shannon's information theory to all of biology. Tom Schneider is best known for inventing sequence logos, a computer graphic depicting patterns in DNA, RNA or protein that is now widely used by molecular biologists. Logos are only the beginning, however, as the information theory measure used to compute them gives results in bits. But why would a binding site have some number of bits? This led to a simple theory: the number of bits in the DNA binding site of a protein is the number needed to find the sites in the genome. Click on the dinosaur to see how these evolve! Next Tom asked how are bits related to binding energy? He solved this problem by using a version of the second law of thermodynamics to convert the bits to the energy needed to select them. Dividing the bits used to define a binding site by the bits that could have been selected for the given energy, he then discovered that the efficiency of DNA binding site selections is near 70% and he constructed a theory to explain this result. Schneider has a number of nanotechnology patents derived in part from this theory. |
|
--- Richard Dawkins, The Blind Watchmaker, 1986, Norton, p. 112. |
|
|
| Note: These pages use two addional windows, a glossary and references. You can use frames in the bar to the left or launch them separately by clicking on these links. Once you have launched the windows, you can instantly look up papers or get glossary definitions. You can try this with the icons to the right in the green bar. |

Sequence logos were invented here!
|
|

Sequence walkers were invented here!
|

Evolution - how DNA gets information!
|

Sphere packing - biological states!
|
Molecular efficiency - a measure of biological states |


 orcid.org/ 0000-0002-9841-1531
orcid.org/ 0000-0002-9841-1531





 How (and why) to find a needle in a haystack
How (and why) to find a needle in a haystack
 Medical Applications of Sequence Walkers
Medical Applications of Sequence Walkers

 frequently
frequently

 Glossary
Glossary



 Lambda Lunch Schedule
Lambda Lunch Schedule weekly
weekly
 The Left handed DNA Hall of Fame has moved.
Please note the
The Left handed DNA Hall of Fame has moved.
Please note the 
 geneff:
Generalizing the Isothermal Efficiency by Using Gaussian Distributions
geneff:
Generalizing the Isothermal Efficiency by Using Gaussian Distributions

 Why Do Restriction Enzymes Prefer 4 and 6 Base DNA Sequences?
Why Do Restriction Enzymes Prefer 4 and 6 Base DNA Sequences?