A Gallery of Sequence Logos
|
Before you make a logo please see:
Recommendations for Making Sequence Logos |
This is a sequence logo of Yeast TATA sites:
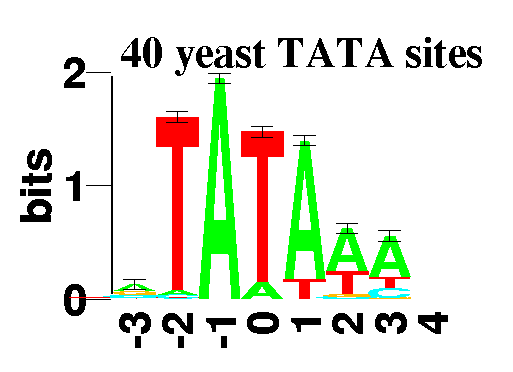
Mark C. Shaner,
Ian M. Blair
and
Thomas D. Schneider
in preparation.
![]()
In the gallery, you can click on each image to get a larger one. The original figures are all in PostScript. Please visit our home page for information about sequence walkers and molecular machines.
 Aligned sequences of Lambda cI and Cro and their logo
(postscript).
This is figure 1 of the paper
Sequence Logos: A Powerful, Yet Simple, Tool.
How to build the figure.
Aligned sequences of Lambda cI and Cro and their logo
(postscript).
This is figure 1 of the paper
Sequence Logos: A Powerful, Yet Simple, Tool.
How to build the figure.
 149 E. coli Ribosome binding sites
(postscript).
This is figure 1 of the paper
Sequence Logos: A New Way to Display
Consensus Sequences
149 E. coli Ribosome binding sites
(postscript).
This is figure 1 of the paper
Sequence Logos: A New Way to Display
Consensus Sequences
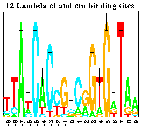 Bacteriophage Lambda cI and Cro
(postscript).
This is figure 2 of the paper
Sequence Logos: A New Way to Display
Consensus Sequences
Bacteriophage Lambda cI and Cro
(postscript).
This is figure 2 of the paper
Sequence Logos: A New Way to Display
Consensus Sequences
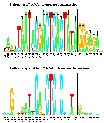 Bacteriophage T7 RNA Polymerase Promoters
(postscript).
This is figure 3 of the paper
Sequence Logos: A New Way to Display
Consensus Sequences
Bacteriophage T7 RNA Polymerase Promoters
(postscript).
This is figure 3 of the paper
Sequence Logos: A New Way to Display
Consensus Sequences
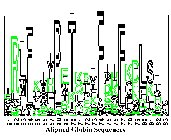
 Globin sequence logo
(postscript).
(postscript for different colors)
This is figure 4 of the paper
Sequence Logos: A New Way to Display
Consensus Sequences.
Globin sequence logo
(postscript).
(postscript for different colors)
This is figure 4 of the paper
Sequence Logos: A New Way to Display
Consensus Sequences.
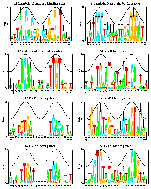 8 E. coli sequence logos
(postscript).
This is figure 6 of the paper
Information Analysis of Sequences that Bind
the Replication Initiator RepA
Papp, P. P., D. K. Chattoraj, and T. D. Schneider. 1993.
J. Mol. Biol. 233: 219-230.
8 E. coli sequence logos
(postscript).
This is figure 6 of the paper
Information Analysis of Sequences that Bind
the Replication Initiator RepA
Papp, P. P., D. K. Chattoraj, and T. D. Schneider. 1993.
J. Mol. Biol. 233: 219-230.
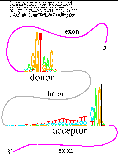 Human Splice Junctions
(postscript).
Information about the dataset used to create these logos.
This is related to figure 1 of the paper
Features of spliceosome evolution and
function inferred from an analysis of the information at human splice
sites
Human Splice Junctions
(postscript).
Information about the dataset used to create these logos.
This is related to figure 1 of the paper
Features of spliceosome evolution and
function inferred from an analysis of the information at human splice
sites
 1055 E. coli Ribosome Binding Sites
(postscript).
1055 E. coli Ribosome Binding Sites
(postscript).
 Partial color aligned listing of the sites.
(postscript for the complete aligned listing)
Partial color aligned listing of the sites.
(postscript for the complete aligned listing)
ASCII aligned listing of the sites.
These sites
were reported in
K. E. Rudd and T. D. Schneider,
Compilation of E. coli Ribosome Binding Sites,
17.19-17.45,
Jeffrey H. Miller,
A Short Course in Bacterial Genetics: A Laboratory
Manual and Handbook for Escherichia coli and Related Bacteria,
Cold Spring Harbor Laboratory Press,
Cold Spring Harbor, New York,
1992.
Information about the data set is here
here.
![]()
Some Online Papers about Sequence Logos:
Sequence Logos: A New Way to Display Consensus Sequences
Sequence Logos: A Powerful, Yet Simple, Tool
![]()
Make Your Own Sequence Logos on the Web:
With Steve Brenner's WebLogo you can create your own sequence logo - just supply a Fasta format file.
![]()
CorreLogo: An online server for 3D sequence logos of RNA and DNA alignments

|
E. Bindewald, T. D. Schneider and B. A. Shapiro, CorreLogo: An online server for 3D sequence logos of RNA and DNA alignments, Nucleic Acids Res. 34, w405-w411, 2006. |
![]()
Logoaid, a Unix script for making sequence logos:
Shmuel Pietrokovski (pietro@sparky.fhcrc.org Fred Hutchinson Cancer Research Center Seattle WA 98104, USA fax: (206) 667 6497; He was formerly at the Structural Biology Department, Weizmann Institute of Science, 76100 Rehovot - ISRAEL fax: 972 (8) 344105 tel: 972 (8) 343367 bppietro@dapsas1.weizmann.ac.il) has made a Unix shell script for creating sequence logos. The source code is available by anonymous ftp from the US or Israel. The Biological Computing Division at the Weizmann Institute has made an example of sequence logo and the files used to create it. The examples are under the "Silicon Graphics software" on the Unix computer used by the biology departments at the Weizmann Institute in Israel.
Here are some comments by
Shmuel Pietrokovski ! pietro@sparky.fhcrc.org
Fred Hutchinson Cancer Research Center ! fax: ##1 (206) 667 5889
1124 Columbia St. Seattle, WA 98104 USA ! tel: ##1 (206) 667 4509
on this technique for looking at protein components:
"We implemented among other things a logo representation for blocks I think its great. You can check it out by choosing the Getblock option, finding some block family you like using the full indexed keyword search, returning to the getblock screen and entering that block family number in the window. There you would see the logo options."
"Each block family has a code(BL#####, # is a digit) with no special significance except being derived from the prosite family number (PS#####)."
"Each block in a block family has logo representation. In order to see a logo you first have to choose a family and then a block in it. If you don't know the family code you can use the "full indexed keyword search of Blocks Database" to find the block using keywords (globin, serine kinase, histone etc). Even if you just want to see any logo you first have to choose a block. You can try any random number but it might happen that there is no such block family. BL00027, BL00622 are some family code I happen to remember, but you might want to see a logo of a specific family (BL01033 is the globin family)."
"We described the logo option in a manuscript about the server. You can FTP it from ftp://howard.fhcrc.org/papers/BLOCKMAKER.ps.
The paper was published at
@article{Henikoff-Pietrokovski1995,
author = "S. Henikoff
and J. G. Henikoff
and W. J. Alford
and S. Pietrokovski",
title = "Automated construction and graphical presentation of protein blocks
from unaligned sequences",
journal = "Gene",
volume = "163",
pages = "GC17-GC26",
year = "1995"}
To see the example of the globin family, go here.
J. Gorodkin, L. J. Heyer, S. Brunak and G. D. Stormo. Displaying the information
contents of structural RNA alignments: the structure logos. Comput. Appl. Biosci.,
Vol. 13, no. 6 pp 583-586, 1997.
This webpage is an attempt to compile ligand sequences for SH3 domains of
various proteins. It will feature the actual alignment, logos, motif, and structural
complexes of binding peptide sequences isolated from combinatorial libraries.
Maintained by:
![]()
RNA Structure Logos
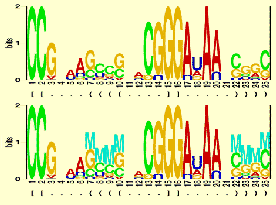 "The structure logo is an extension of the sequence logo by Schneider and
Stephens. We have extended the standard sequence logo to cope with any prior
nucleotide distribution as well as allowing for gaps in the alignments, and
indicate mutual information of basepaired positions in RNA. Thus the logo is
composed by a sequence part and a structure/basepair part."
"The structure logo is an extension of the sequence logo by Schneider and
Stephens. We have extended the standard sequence logo to cope with any prior
nucleotide distribution as well as allowing for gaps in the alignments, and
indicate mutual information of basepaired positions in RNA. Thus the logo is
composed by a sequence part and a structure/basepair part."
![]()
Sequence Logos of SH3 Domain Ligands
Department of Biology
University of North Carolina
Chapel Hill, NC 27599-3280
Sorry - as of 2002 Feb 12 the link to http://kay02.bio.unc.edu/sh3.html is broken. Brian Kay has disappeared ... Do you know where he went?
![]()
Cleavage site analysis in picornaviral polyproteins: Discovering cellular targets by neural networks
Nikolaj BLOM, Jan HANSEN, Dieter BLAAS, and Soren Brunak,Protein Science (1996), 5: 2203- 2216.
Sequence Logos were used to study protein structures determined by neural nets. The original figures were in color. The ones on the web are black and white and low quality.
![]() Logo Sequence Conservation in Ornithine
Transcarbamylase (OTC)
Logo Sequence Conservation in Ornithine
Transcarbamylase (OTC)
M. Tuchman, H. Morizono, B.S. Rajagopal, R.J. Plante, and N. M.
Allewell,
The Biochemical and Molecular Spectrum of Ornithine Transcarbamylase
Deficiency,
J. Inher. Metab. Dis. 21 (Suppl 1) 1998 40-58.
![]() Prediction Tools for Protein Homology Domain-Associated
Post-Translational Modifications in the RESID Database
J.S. Garavelli, D.J. Miller, and G.Y. Srinivasarao,
(Protein Information Resource,
National Biomedical Research Foundation,
Georgetown University Medical Center, Washington, DC 20007,
Poster presented at the Protein Society Meeting, July 23 - 27, 1999, Boston, MA)
Prediction Tools for Protein Homology Domain-Associated
Post-Translational Modifications in the RESID Database
J.S. Garavelli, D.J. Miller, and G.Y. Srinivasarao,
(Protein Information Resource,
National Biomedical Research Foundation,
Georgetown University Medical Center, Washington, DC 20007,
Poster presented at the Protein Society Meeting, July 23 - 27, 1999, Boston, MA)
![]()
 as of 2000 Jan 4
as of 2000 Jan 4
Sequence Logos in George Church's Course
- Biophysic101: Genomics and Computational Biology
- Church Lab WWW Server
- Human RNA-splice
- Protein starts
- Overview
- 8 methods for predicting regulons
- AlignACE Example:
- AlignACE Example:
- AlignACE Example:
- AlignACE Example: Final Results
- Church Lab WWW Server
Sequence Logos in Excel
 as of 2000 July 27
as of 2000 July 27
@article{Delamarche2000,
author = "C. Delamarche",
title = "{Color and graphic display (CGD): programs for multiple sequence
alignment analysis in spreadsheet software}",
journal = "Biotechniques",
volume = "29",
pages = "100-4, 106-7",
year = "2000"}
The program generates logos in Excel.
The software is supposed to be
in the software library at
BioTechniques
but I could not find it as of 2000 July 27.
These logos do not have the letters sorted
to put the most frequent letter at the top, but I have
written to Christian Delamarche
(cdelam@univ-rennes1.fr) to suggest this.
![]()
Sequence Logos for Escherichia coli Transcription Factor Binding Sites
 as of 2001 March 27
as of 2001 March 27
- Database
- reference: Escherichia coli Transcription Factor Binding Sites McCue, L.A., Thompson, W., Carmack, C.S., Ryan, M.P., Liu, J.S., Derbyshire, V. and Lawrence, C.E. Phylogenetic footprinting of transcription factor binding sites in proteobacterial genomes NAR 2001 29:774-782. PDF
- Environmentally induced foregut remodeling by PHA-4/FoxA and DAF-12/NHR Science. 2004 Sep 17;305(5691):1743-1746. They used sequence logos to show binding sites.

Human Sequence Logo. From the left, it's: Ryan (G), Karen (A), Denise (T), and Elaine (C). From Karen Lewis 1998.
![]()

Schneider Lab
origin: before 1996 June 12
updated: 2015 Jan 15
![]()